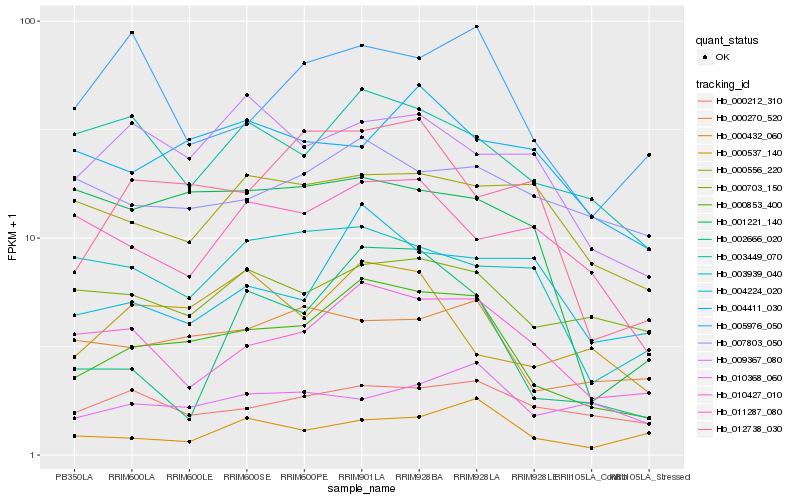
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_400 |
0.0 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 2 |
Hb_010427_010 |
0.1471069205 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 3 |
Hb_000270_520 |
0.155651144 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_012738_030 |
0.1655118353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003449_070 |
0.1680610197 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 6 |
Hb_005976_050 |
0.1754115105 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 7 |
Hb_000212_310 |
0.1756624688 |
- |
- |
hypothetical protein JCGZ_24350 [Jatropha curcas] |
| 8 |
Hb_000703_150 |
0.1779672977 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A [Jatropha curcas] |
| 9 |
Hb_004224_020 |
0.1780492322 |
- |
- |
PREDICTED: uncharacterized protein LOC105643786 [Jatropha curcas] |
| 10 |
Hb_009367_080 |
0.1811597313 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 42 [Jatropha curcas] |
| 11 |
Hb_003939_040 |
0.1822728479 |
- |
- |
hypothetical protein CICLE_v10014350mg [Citrus clementina] |
| 12 |
Hb_002666_020 |
0.1826171971 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 13 |
Hb_000537_140 |
0.1855439307 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 14 |
Hb_001221_140 |
0.1861177036 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000432_060 |
0.1879947088 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_004411_030 |
0.1887801103 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 17 |
Hb_011287_080 |
0.1895114788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010368_060 |
0.1898443047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007803_050 |
0.1908079001 |
- |
- |
PREDICTED: uncharacterized protein LOC105129170 isoform X2 [Populus euphratica] |
| 20 |
Hb_000556_220 |
0.1914020625 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |