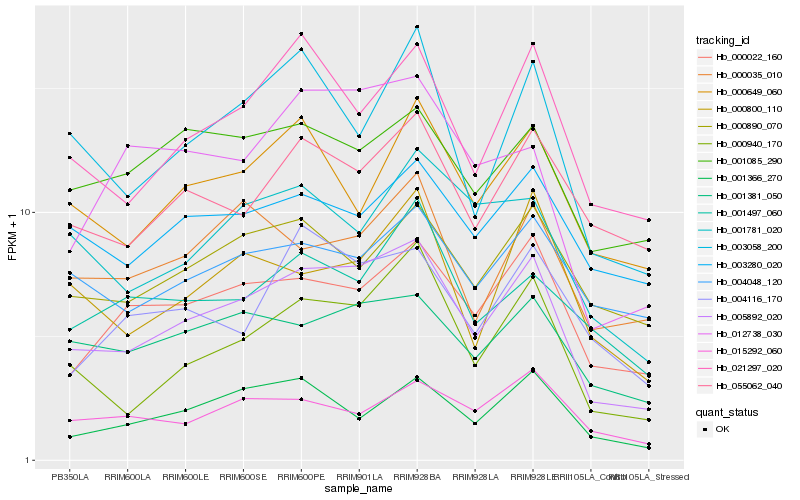
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005892_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632170 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000940_170 |
0.1015145035 |
- |
- |
hypothetical protein VITISV_029834 [Vitis vinifera] |
| 3 |
Hb_021297_020 |
0.1122123053 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 4 |
Hb_000022_160 |
0.1151456219 |
- |
- |
- |
| 5 |
Hb_001381_050 |
0.1164803752 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_003058_200 |
0.1217633709 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 7 |
Hb_012738_030 |
0.129565948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001366_270 |
0.130021194 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 9 |
Hb_000890_070 |
0.1316697244 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_055062_040 |
0.1324999985 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 11 |
Hb_000800_110 |
0.1329778344 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000035_010 |
0.1344812463 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001781_020 |
0.1362924462 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 14 |
Hb_001085_290 |
0.1363777436 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 15 |
Hb_000649_060 |
0.1369762932 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 16 |
Hb_004116_170 |
0.1373011453 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003280_020 |
0.1386496479 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_015292_060 |
0.1388608158 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 19 |
Hb_004048_120 |
0.1395941461 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 20 |
Hb_001497_060 |
0.1396924687 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |