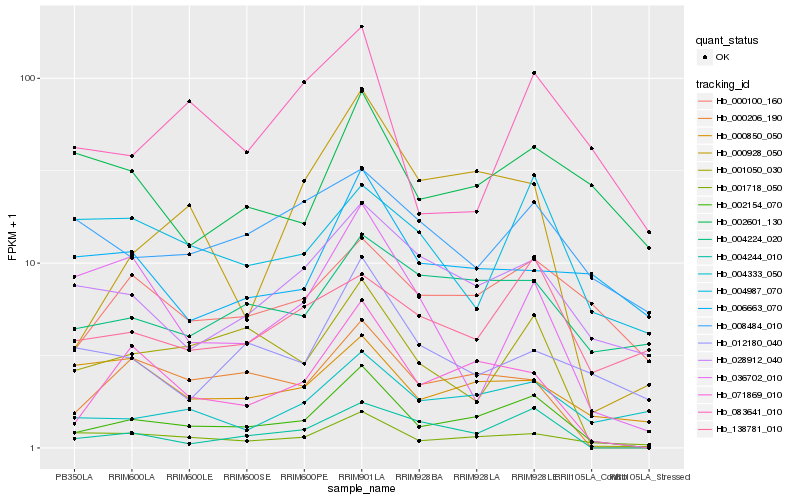
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002154_070 |
0.0 |
desease resistance |
Gene Name: AAA_25 |
hypothetical protein Csa_6G014750 [Cucumis sativus] |
| 2 |
Hb_071869_010 |
0.1590418177 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001718_050 |
0.188219207 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Brassica rapa] |
| 4 |
Hb_004244_010 |
0.1894752081 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 5 |
Hb_000206_190 |
0.1923111 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 6 |
Hb_001050_030 |
0.1959164983 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 7 |
Hb_028912_040 |
0.2135211976 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 8 |
Hb_000928_050 |
0.2139255647 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 9 |
Hb_000100_160 |
0.226844174 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 10 |
Hb_004333_050 |
0.2289288815 |
- |
- |
- |
| 11 |
Hb_004224_020 |
0.2319377229 |
- |
- |
PREDICTED: uncharacterized protein LOC105643786 [Jatropha curcas] |
| 12 |
Hb_012180_040 |
0.2364276637 |
- |
- |
hypothetical protein VITISV_042517 [Vitis vinifera] |
| 13 |
Hb_083641_010 |
0.236646467 |
- |
- |
SECY isoform 1 [Theobroma cacao] |
| 14 |
Hb_000850_050 |
0.2442476275 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 15 |
Hb_138781_010 |
0.2458171281 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 16 |
Hb_002601_130 |
0.2463066067 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 17 |
Hb_036702_010 |
0.2469473314 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 18 |
Hb_008484_010 |
0.2498768588 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 19 |
Hb_004987_070 |
0.250459376 |
- |
- |
PREDICTED: heat shock 70 kDa protein 17-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_006663_070 |
0.251164777 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |