| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
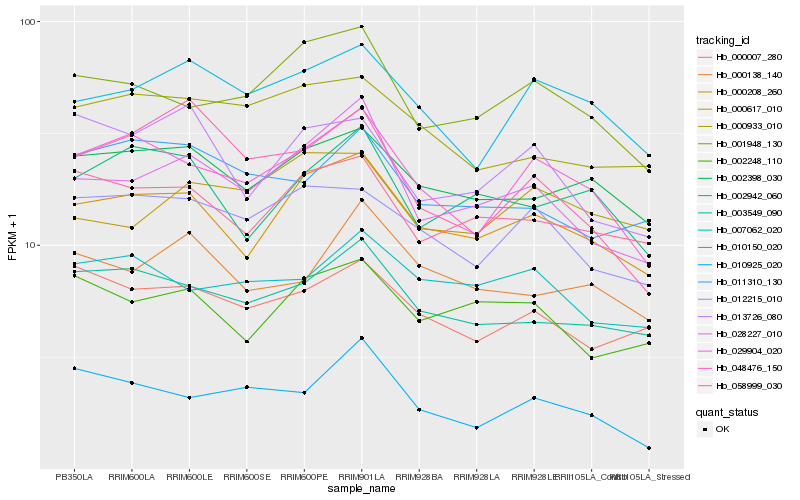
Hb_028227_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 2 |
Hb_000208_260 |
0.0837844804 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_001948_130 |
0.0905633165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 4 |
Hb_003549_090 |
0.1033987329 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 5 |
Hb_002248_110 |
0.1047728766 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 6 |
Hb_058999_030 |
0.1076912931 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_029904_020 |
0.1086990449 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 8 |
Hb_000138_140 |
0.1168634874 |
- |
- |
hypothetical protein B456_012G1014001, partial [Gossypium raimondii] |
| 9 |
Hb_002398_030 |
0.1188386885 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_010150_020 |
0.1191366738 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 11 |
Hb_000007_280 |
0.1206485532 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 12 |
Hb_048476_150 |
0.1211941147 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_012215_010 |
0.1216249016 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 14 |
Hb_000933_010 |
0.1218148126 |
- |
- |
- |
| 15 |
Hb_010925_020 |
0.1222723619 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 16 |
Hb_011310_130 |
0.1230323688 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_002942_060 |
0.1231253491 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 18 |
Hb_000617_010 |
0.123182143 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 19 |
Hb_007062_020 |
0.1247262372 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 20 |
Hb_013726_080 |
0.1249707667 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |