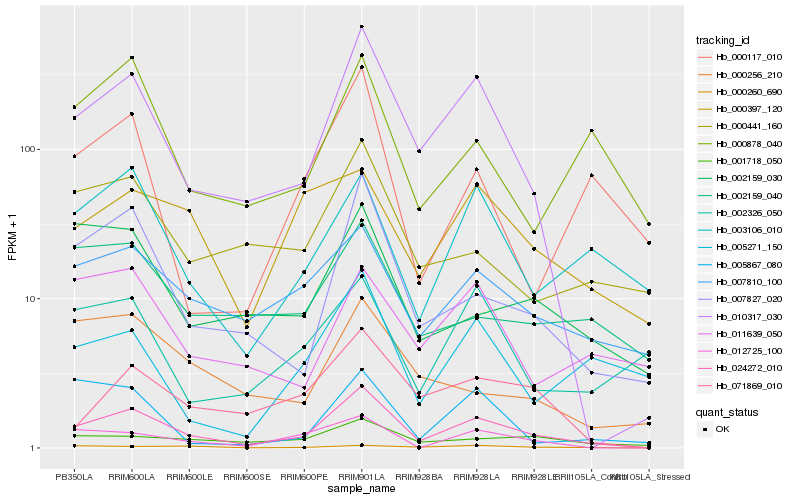
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024272_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 2 |
Hb_010317_030 |
0.1419369734 |
- |
- |
unknown [Picea sitchensis] |
| 3 |
Hb_007827_020 |
0.1826098384 |
- |
- |
- |
| 4 |
Hb_007810_100 |
0.211686733 |
- |
- |
Synaptic glycoprotein SC2, putative [Ricinus communis] |
| 5 |
Hb_071869_010 |
0.2136639634 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 6 |
Hb_012725_100 |
0.2180794938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003106_010 |
0.2240803714 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 8 |
Hb_001718_050 |
0.2329921723 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Brassica rapa] |
| 9 |
Hb_000441_160 |
0.2341609921 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 10 |
Hb_002159_030 |
0.2380392925 |
- |
- |
AP-1 complex subunit gamma-2, putative [Ricinus communis] |
| 11 |
Hb_000878_040 |
0.2395344712 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000397_120 |
0.2430860838 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 13 |
Hb_005271_150 |
0.2438953676 |
- |
- |
PREDICTED: EKC/KEOPS complex subunit Tprkb-like [Malus domestica] |
| 14 |
Hb_000117_010 |
0.2460761492 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like [Malus domestica] |
| 15 |
Hb_000260_690 |
0.2462426985 |
- |
- |
disease resistance protein (CC-NBS-LRR class) family protein [Medicago truncatula] |
| 16 |
Hb_002159_040 |
0.2465687221 |
- |
- |
hypothetical protein PRUPE_ppa001563mg [Prunus persica] |
| 17 |
Hb_000256_210 |
0.247496976 |
- |
- |
PREDICTED: 40S ribosomal protein S11-like [Phoenix dactylifera] |
| 18 |
Hb_005867_080 |
0.2476410103 |
- |
- |
PREDICTED: protein NETWORKED 2A [Jatropha curcas] |
| 19 |
Hb_011639_050 |
0.249435059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002326_050 |
0.2495782743 |
- |
- |
PREDICTED: uncharacterized protein LOC105633701 [Jatropha curcas] |