| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
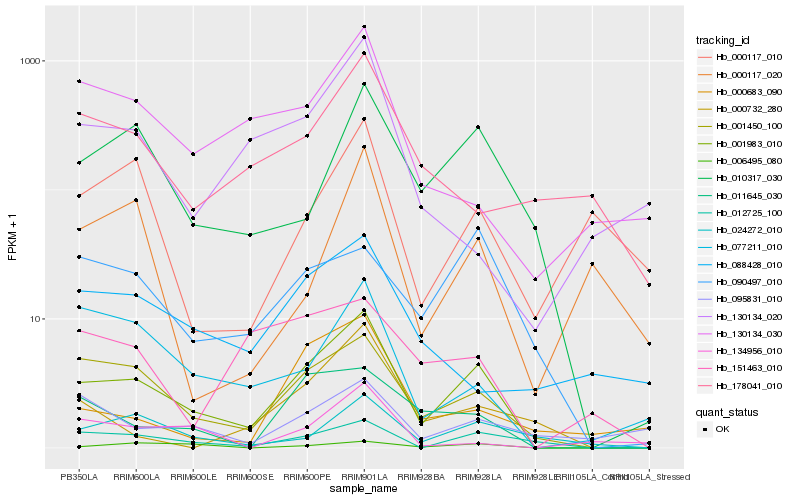
Hb_001983_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001450_100 |
0.1897740558 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 3 |
Hb_012725_100 |
0.2389652114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010317_030 |
0.2429499896 |
- |
- |
unknown [Picea sitchensis] |
| 5 |
Hb_011645_030 |
0.2581152392 |
- |
- |
- |
| 6 |
Hb_024272_010 |
0.2613909593 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 7 |
Hb_077211_010 |
0.2727390598 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Jatropha curcas] |
| 8 |
Hb_006495_080 |
0.2751572833 |
- |
- |
hypothetical protein PRUPE_ppb025438mg, partial [Prunus persica] |
| 9 |
Hb_090497_010 |
0.2815462216 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 10 |
Hb_000732_280 |
0.2826707139 |
- |
- |
hypothetical protein B456_009G173700 [Gossypium raimondii] |
| 11 |
Hb_130134_030 |
0.2863972761 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 12 |
Hb_000683_090 |
0.2920295673 |
- |
- |
stress enhanced protein [Medicago truncatula] |
| 13 |
Hb_088428_010 |
0.294803606 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 14 |
Hb_130134_020 |
0.3007395498 |
- |
- |
- |
| 15 |
Hb_178041_010 |
0.3012698813 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Populus euphratica] |
| 16 |
Hb_151463_010 |
0.3014638764 |
- |
- |
hypothetical protein B456_004G013100 [Gossypium raimondii] |
| 17 |
Hb_095831_010 |
0.3041098506 |
- |
- |
PREDICTED: uncharacterized protein LOC104884649 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_000117_010 |
0.3046824161 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like [Malus domestica] |
| 19 |
Hb_134956_010 |
0.305770769 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Jatropha curcas] |
| 20 |
Hb_000117_020 |
0.3068600169 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |