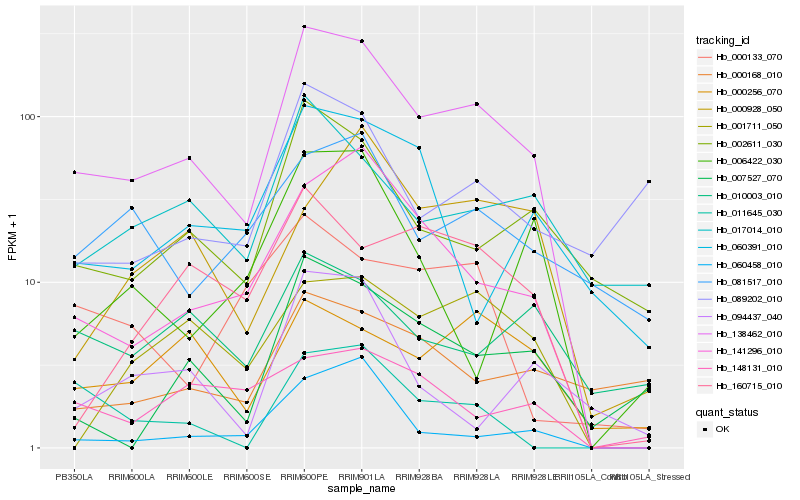
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138462_010 |
0.0 |
- |
- |
PREDICTED: SUMO-conjugating enzyme UBC9-A-like isoform X1 [Elaeis guineensis] |
| 2 |
Hb_141296_010 |
0.1868421197 |
- |
- |
- |
| 3 |
Hb_002611_030 |
0.2066559431 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 4 |
Hb_017014_010 |
0.2239344063 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_060458_010 |
0.2272171197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001711_050 |
0.2316745069 |
- |
- |
- |
| 7 |
Hb_007527_070 |
0.2345929459 |
- |
- |
PREDICTED: uncharacterized protein LOC105637197 [Jatropha curcas] |
| 8 |
Hb_000256_070 |
0.2386637592 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial [Jatropha curcas] |
| 9 |
Hb_160715_010 |
0.2395893936 |
- |
- |
- |
| 10 |
Hb_081517_010 |
0.2460243682 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 11 |
Hb_060391_010 |
0.2495726734 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_011645_030 |
0.2548813501 |
- |
- |
- |
| 13 |
Hb_148131_010 |
0.256803965 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12-like, partial [Jatropha curcas] |
| 14 |
Hb_010003_010 |
0.2574613272 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000928_050 |
0.2591843117 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 16 |
Hb_094437_040 |
0.260274365 |
- |
- |
ankyrin repeat family protein, partial [Manihot esculenta] |
| 17 |
Hb_000168_010 |
0.2620978135 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 18 |
Hb_006422_030 |
0.2626285662 |
- |
- |
- |
| 19 |
Hb_089202_010 |
0.2628993492 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 20 |
Hb_000133_070 |
0.2637791127 |
- |
- |
- |