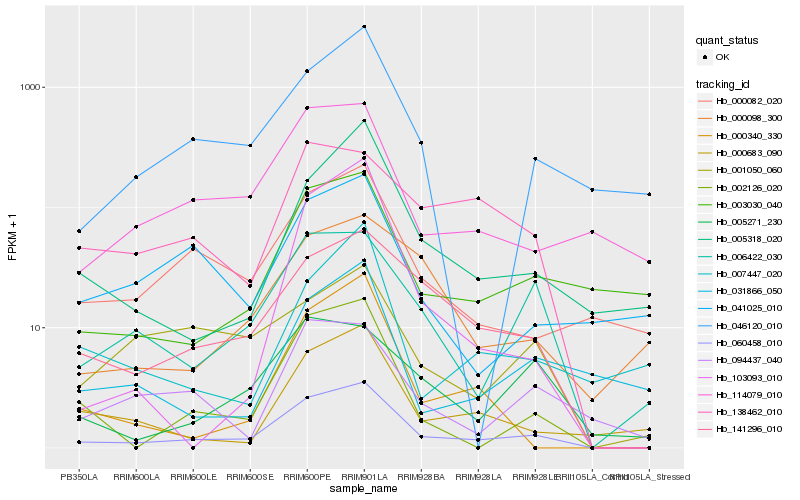
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060458_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_141296_010 |
0.1702009413 |
- |
- |
- |
| 3 |
Hb_002126_020 |
0.1890963735 |
- |
- |
ferrochelatase [Medicago truncatula] |
| 4 |
Hb_006422_030 |
0.1935818573 |
- |
- |
- |
| 5 |
Hb_000082_020 |
0.202290442 |
- |
- |
- |
| 6 |
Hb_005318_020 |
0.2067239446 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_114079_010 |
0.2067878008 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 8 |
Hb_003030_040 |
0.2105446014 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 9 |
Hb_000340_330 |
0.2129740243 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 10 |
Hb_000683_090 |
0.2151968133 |
- |
- |
stress enhanced protein [Medicago truncatula] |
| 11 |
Hb_046120_010 |
0.2222194955 |
- |
- |
- |
| 12 |
Hb_138462_010 |
0.2272171197 |
- |
- |
PREDICTED: SUMO-conjugating enzyme UBC9-A-like isoform X1 [Elaeis guineensis] |
| 13 |
Hb_094437_040 |
0.2291306775 |
- |
- |
ankyrin repeat family protein, partial [Manihot esculenta] |
| 14 |
Hb_005271_230 |
0.2310107725 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 15 |
Hb_000098_300 |
0.2359474732 |
- |
- |
- |
| 16 |
Hb_103093_010 |
0.2379494711 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 17 |
Hb_031866_050 |
0.2393643898 |
- |
- |
PREDICTED: uncharacterized protein LOC101756357 [Setaria italica] |
| 18 |
Hb_001050_060 |
0.2404518672 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 19 |
Hb_041025_010 |
0.242008656 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 20 |
Hb_007447_020 |
0.2438965136 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |