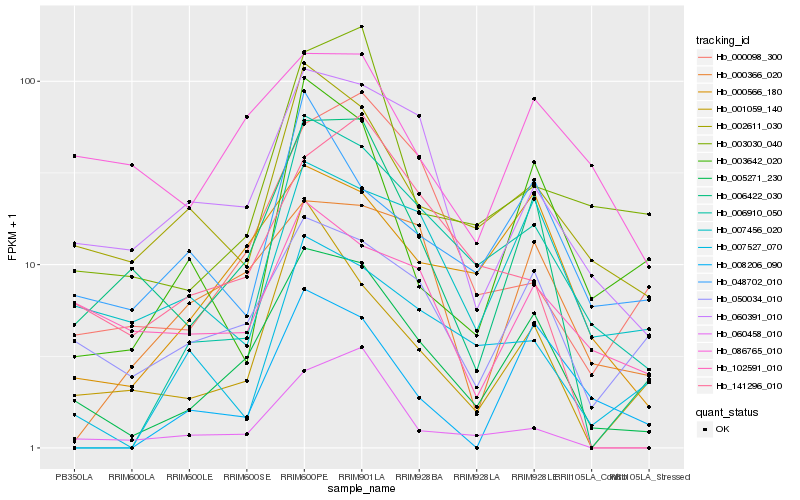
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 2 |
Hb_006422_030 |
0.1516336984 |
- |
- |
- |
| 3 |
Hb_000566_180 |
0.1732906062 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 4 |
Hb_050034_010 |
0.182940945 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 5 |
Hb_060391_010 |
0.1907040128 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_006910_050 |
0.1976769451 |
- |
- |
hypothetical protein CISIN_1g0480481mg, partial [Citrus sinensis] |
| 7 |
Hb_002611_030 |
0.2017013598 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 8 |
Hb_007456_020 |
0.209064296 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_000366_020 |
0.2220057011 |
- |
- |
- |
| 10 |
Hb_001059_140 |
0.2276027728 |
- |
- |
hypothetical protein POPTR_0004s03390g [Populus trichocarpa] |
| 11 |
Hb_003642_020 |
0.2305781769 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 12 |
Hb_060458_010 |
0.2310107725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_086765_010 |
0.23467757 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 14 |
Hb_102591_010 |
0.234894307 |
- |
- |
hypothetical protein JCGZ_09876 [Jatropha curcas] |
| 15 |
Hb_007527_070 |
0.2359683408 |
- |
- |
PREDICTED: uncharacterized protein LOC105637197 [Jatropha curcas] |
| 16 |
Hb_000098_300 |
0.2360954914 |
- |
- |
- |
| 17 |
Hb_048702_010 |
0.2375785903 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_003030_040 |
0.2382380113 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 19 |
Hb_008206_090 |
0.2419411054 |
- |
- |
PREDICTED: uncharacterized protein LOC100811033 [Glycine max] |
| 20 |
Hb_141296_010 |
0.2421099119 |
- |
- |
- |