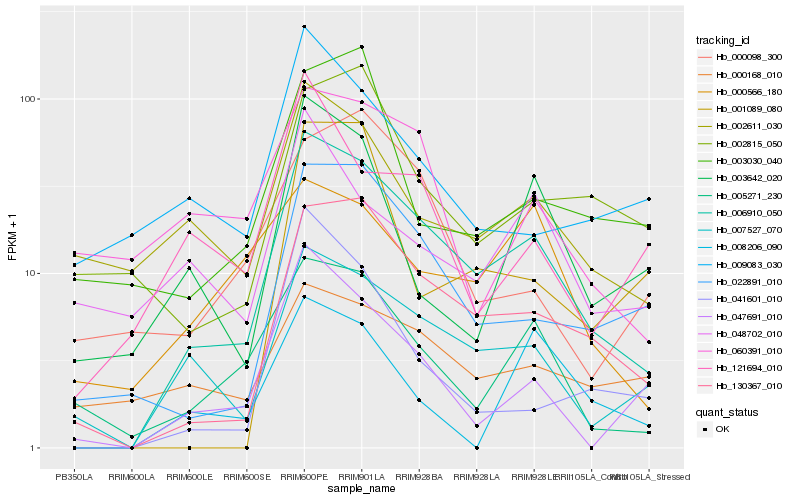
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006910_050 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0480481mg, partial [Citrus sinensis] |
| 2 |
Hb_130367_010 |
0.1431019289 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 3 |
Hb_007527_070 |
0.1680815229 |
- |
- |
PREDICTED: uncharacterized protein LOC105637197 [Jatropha curcas] |
| 4 |
Hb_022891_010 |
0.197550632 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 5 |
Hb_005271_230 |
0.1976769451 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 6 |
Hb_047691_010 |
0.2170320464 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |
| 7 |
Hb_002611_030 |
0.2292277238 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 8 |
Hb_003642_020 |
0.2431649752 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 9 |
Hb_041601_010 |
0.2441918511 |
- |
- |
- |
| 10 |
Hb_002815_050 |
0.2457627759 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 11 |
Hb_000566_180 |
0.2481995892 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 12 |
Hb_000168_010 |
0.2489485497 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 13 |
Hb_121694_010 |
0.2495198817 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 14 |
Hb_001089_080 |
0.2507661174 |
- |
- |
hypothetical protein glysoja_044047 [Glycine soja] |
| 15 |
Hb_060391_010 |
0.2519137422 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_009083_030 |
0.2526460496 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 17 |
Hb_000098_300 |
0.2538649789 |
- |
- |
- |
| 18 |
Hb_008206_090 |
0.2538946696 |
- |
- |
PREDICTED: uncharacterized protein LOC100811033 [Glycine max] |
| 19 |
Hb_003030_040 |
0.254651615 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 20 |
Hb_048702_010 |
0.2555585901 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |