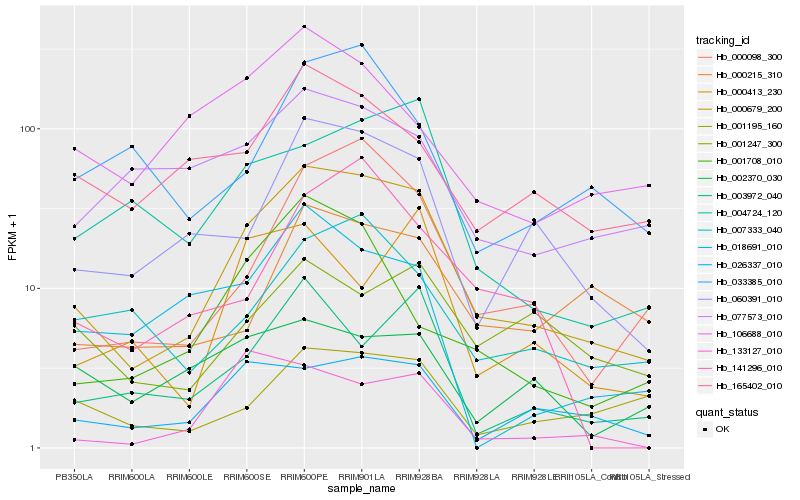
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_200 |
0.0 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 2 |
Hb_060391_010 |
0.1765105398 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000098_300 |
0.1768399973 |
- |
- |
- |
| 4 |
Hb_026337_010 |
0.1939962791 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10002052mg [Citrus clementina] |
| 5 |
Hb_007333_040 |
0.2073417189 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 6 |
Hb_004724_120 |
0.2101140351 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 7 |
Hb_003972_040 |
0.210972578 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 8 |
Hb_165402_010 |
0.2125505006 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 9 |
Hb_000215_310 |
0.2145771907 |
- |
- |
- |
| 10 |
Hb_001195_160 |
0.2148789566 |
- |
- |
- |
| 11 |
Hb_001708_010 |
0.2199194434 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Malus domestica] |
| 12 |
Hb_077573_010 |
0.2214108583 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_033385_010 |
0.227753815 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 14 |
Hb_141296_010 |
0.2296525657 |
- |
- |
- |
| 15 |
Hb_106688_010 |
0.2304018746 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 16 |
Hb_001247_300 |
0.2322866901 |
- |
- |
hypothetical protein JCGZ_23167 [Jatropha curcas] |
| 17 |
Hb_000413_230 |
0.2341396666 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 18 |
Hb_018691_010 |
0.2359140744 |
- |
- |
- |
| 19 |
Hb_133127_010 |
0.2391398957 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 20 |
Hb_002370_030 |
0.2414218342 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |