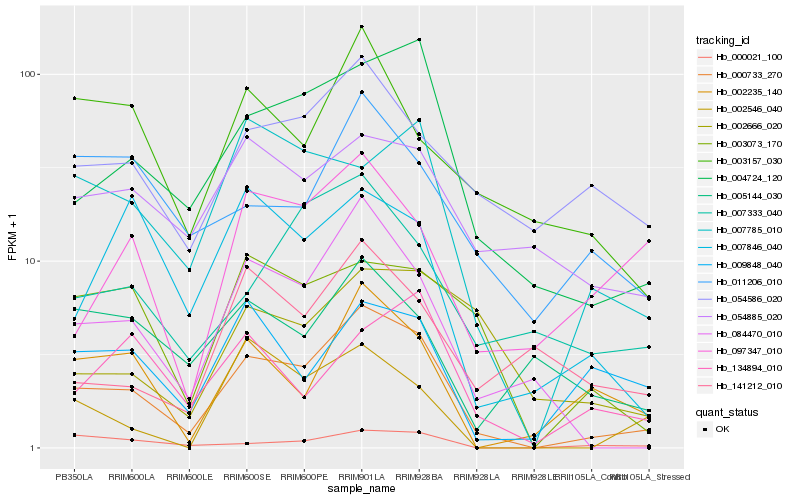
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000733_270 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-alpha catalytic subunit [Dasypus novemcinctus] |
| 2 |
Hb_004724_120 |
0.177600916 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 3 |
Hb_084470_010 |
0.1850271718 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 4 |
Hb_002235_140 |
0.2214105367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000021_100 |
0.2262760881 |
- |
- |
- |
| 6 |
Hb_007785_010 |
0.2283418541 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 7 |
Hb_003073_170 |
0.2317897789 |
- |
- |
- |
| 8 |
Hb_007333_040 |
0.2338340294 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 9 |
Hb_054586_020 |
0.2353196435 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 10 |
Hb_009848_040 |
0.2387193124 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 11 |
Hb_054885_020 |
0.2422987786 |
- |
- |
- |
| 12 |
Hb_003157_030 |
0.2431851783 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 13 |
Hb_011206_010 |
0.2477787403 |
- |
- |
PREDICTED: something about silencing protein 10 isoform X2 [Vitis vinifera] |
| 14 |
Hb_007846_040 |
0.2484014691 |
- |
- |
PREDICTED: uncharacterized protein LOC105646691 [Jatropha curcas] |
| 15 |
Hb_134894_010 |
0.2503264245 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_141212_010 |
0.2526598882 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 17 |
Hb_097347_010 |
0.2536208757 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like, partial [Malus domestica] |
| 18 |
Hb_005144_030 |
0.2542751123 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 19 |
Hb_002666_020 |
0.2547815058 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 20 |
Hb_002546_040 |
0.2560858876 |
- |
- |
hypothetical protein CISIN_1g005599mg [Citrus sinensis] |