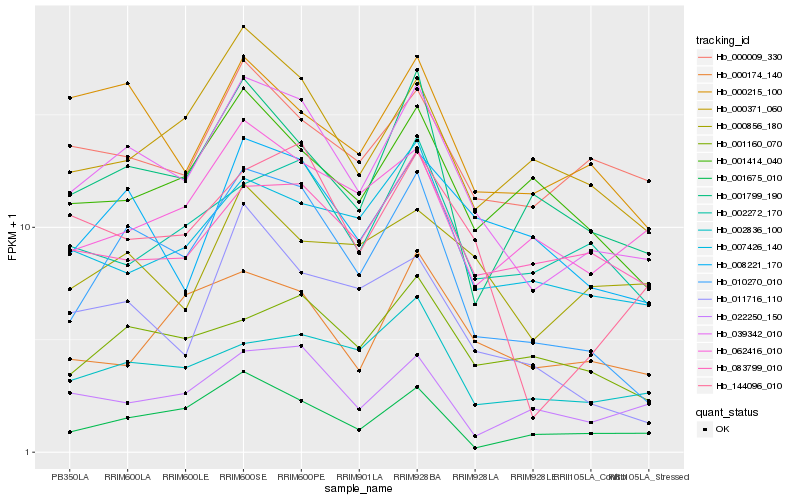
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039342_010 |
0.0 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 2 |
Hb_010270_010 |
0.1038828121 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_144096_010 |
0.1347582118 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 4 |
Hb_008221_170 |
0.1358899023 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 5 |
Hb_000371_060 |
0.1427088245 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 6 |
Hb_007426_140 |
0.1475011517 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 7 |
Hb_000215_100 |
0.1483335988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001799_190 |
0.1489006968 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 9 |
Hb_000009_330 |
0.1489578298 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_002836_100 |
0.1507769155 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002272_170 |
0.1517523208 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 12 |
Hb_001675_010 |
0.1524834729 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 13 |
Hb_011716_110 |
0.1525484738 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 14 |
Hb_083799_010 |
0.1527499573 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_062416_010 |
0.1540489691 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 16 |
Hb_001160_070 |
0.1550904256 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 17 |
Hb_022250_150 |
0.1557019449 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 18 |
Hb_001414_040 |
0.1559049426 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000174_140 |
0.1596408242 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 20 |
Hb_000856_180 |
0.1601128635 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |