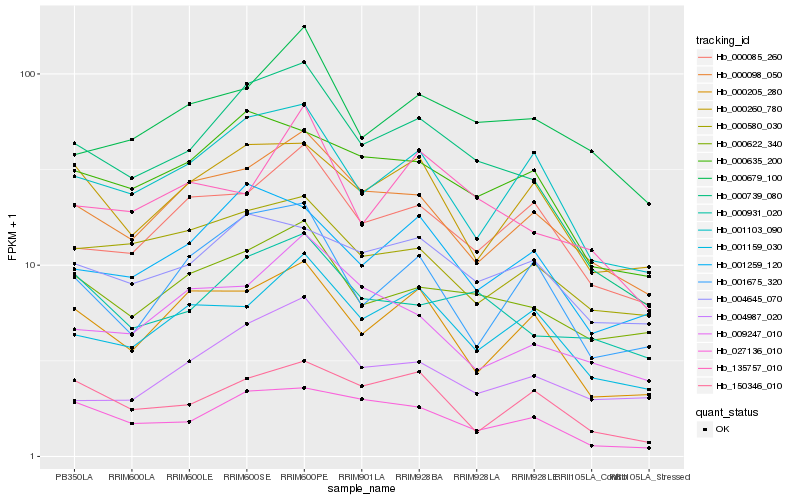
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_080 |
0.0 |
- |
- |
PREDICTED: 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase [Jatropha curcas] |
| 2 |
Hb_027136_010 |
0.1187704844 |
- |
- |
hypothetical protein L484_010426 [Morus notabilis] |
| 3 |
Hb_000635_200 |
0.1264837583 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 4 |
Hb_001103_090 |
0.1272310252 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 5 |
Hb_000098_050 |
0.1289928195 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 6 |
Hb_000931_020 |
0.134795072 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000622_340 |
0.1354729688 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 8 |
Hb_001259_120 |
0.1391463504 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 9 |
Hb_000205_280 |
0.1410140943 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_001159_030 |
0.1419709497 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 11 |
Hb_000085_260 |
0.1454552471 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 12 |
Hb_001675_320 |
0.1473716792 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 13 |
Hb_004645_070 |
0.1477905296 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_000679_100 |
0.149247964 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 15 |
Hb_135757_010 |
0.1497372053 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 16 |
Hb_004987_020 |
0.1516197702 |
- |
- |
- |
| 17 |
Hb_000260_780 |
0.1521858436 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Jatropha curcas] |
| 18 |
Hb_000580_030 |
0.1530343991 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 19 |
Hb_009247_010 |
0.153384065 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_150346_010 |
0.1540619548 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |