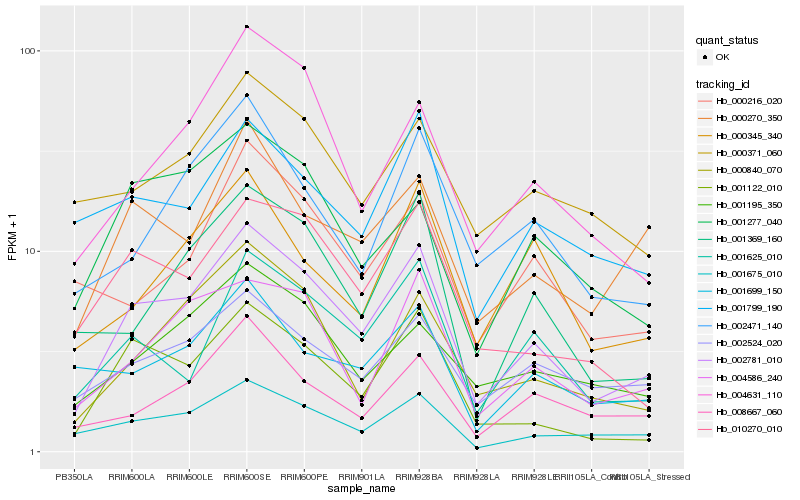
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002781_010 |
0.0 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001675_010 |
0.1236392832 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 3 |
Hb_001122_010 |
0.1295524558 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 4 |
Hb_004586_240 |
0.1375491627 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 5 |
Hb_001625_010 |
0.1380862671 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 6 |
Hb_010270_010 |
0.1396117414 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000840_070 |
0.1404548879 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_001369_160 |
0.1433726914 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 9 |
Hb_002524_020 |
0.1434999508 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 10 |
Hb_000270_350 |
0.1485331683 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 11 |
Hb_004631_110 |
0.1486666057 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_002471_140 |
0.1513884006 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 13 |
Hb_001195_350 |
0.1518684512 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 14 |
Hb_008667_060 |
0.1524843019 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 15 |
Hb_001277_040 |
0.1571823779 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 16 |
Hb_001799_190 |
0.1597641877 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 17 |
Hb_000345_340 |
0.1605595522 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 18 |
Hb_000371_060 |
0.1627761323 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 19 |
Hb_000216_020 |
0.1632838165 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 20 |
Hb_001699_150 |
0.1639096257 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |