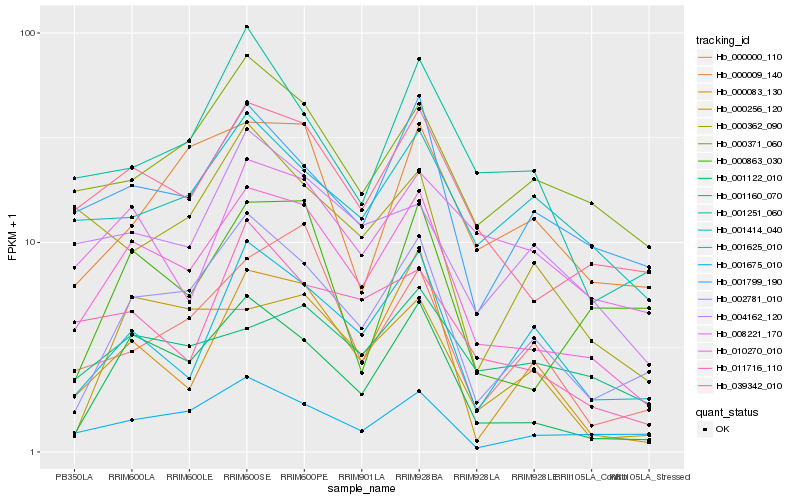
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010270_010 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_039342_010 |
0.1038828121 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 3 |
Hb_001122_010 |
0.1174331074 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 4 |
Hb_002781_010 |
0.1396117414 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001675_010 |
0.1561210665 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 6 |
Hb_001799_190 |
0.161984974 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 7 |
Hb_011716_110 |
0.1621405224 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 8 |
Hb_000371_060 |
0.1663672706 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 9 |
Hb_001625_010 |
0.1669054513 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 10 |
Hb_004162_120 |
0.1724329457 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 11 |
Hb_000083_130 |
0.1729227026 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 12 |
Hb_001251_060 |
0.1747539842 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 13 |
Hb_001160_070 |
0.1782906795 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 14 |
Hb_008221_170 |
0.1808585469 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 15 |
Hb_000009_140 |
0.1816738433 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 16 |
Hb_000000_110 |
0.1817186784 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 17 |
Hb_001414_040 |
0.1819533948 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000362_090 |
0.1828387768 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000256_120 |
0.1856139833 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 20 |
Hb_000863_030 |
0.1856408353 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |