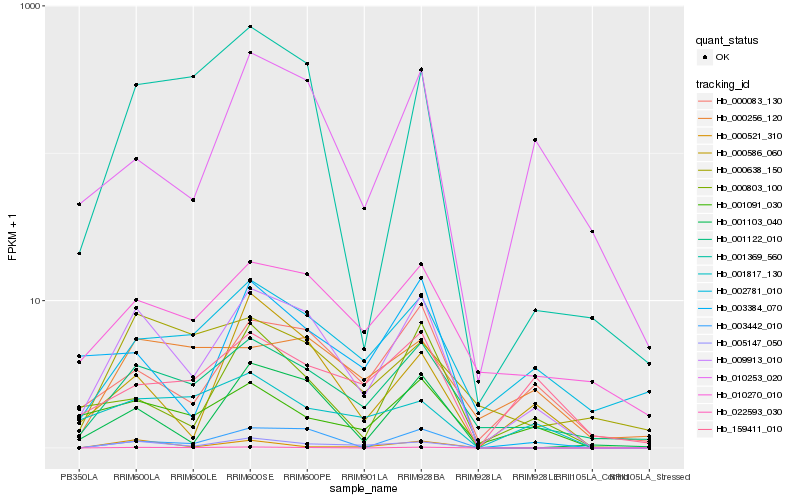
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009913_010 |
0.0 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001122_010 |
0.1693783943 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 3 |
Hb_005147_050 |
0.1888511475 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 4 |
Hb_000083_130 |
0.1976285114 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 5 |
Hb_001103_040 |
0.2105247823 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 6 |
Hb_010270_010 |
0.2205026963 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001091_030 |
0.2231131712 |
- |
- |
hypothetical protein AALP_AA1G173500 [Arabis alpina] |
| 8 |
Hb_001369_560 |
0.2235885005 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 9 |
Hb_022593_030 |
0.2258413958 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 10 |
Hb_000586_060 |
0.2320363963 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 11 |
Hb_010253_020 |
0.2348003735 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_003442_010 |
0.2383226419 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 13 |
Hb_000803_100 |
0.2383549263 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 14 |
Hb_003384_070 |
0.2420838318 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 15 |
Hb_002781_010 |
0.2434457894 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_159411_010 |
0.2497810931 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 17 |
Hb_000256_120 |
0.2579654359 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 18 |
Hb_001817_130 |
0.2588094566 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 isoform X3 [Cucumis melo] |
| 19 |
Hb_000521_310 |
0.259645189 |
- |
- |
- |
| 20 |
Hb_000638_150 |
0.2600562564 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |