| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
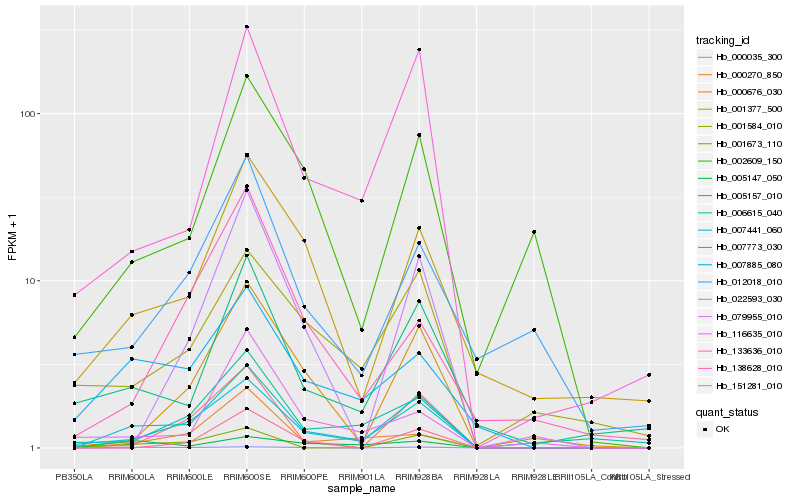
Hb_007441_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_007773_030 |
0.1523562282 |
- |
- |
- |
| 3 |
Hb_007885_080 |
0.1904291533 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000270_850 |
0.1953716059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_133636_010 |
0.2230592706 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 6 |
Hb_001377_500 |
0.2265275352 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 7 |
Hb_000676_030 |
0.2271291661 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 8 |
Hb_005157_010 |
0.2272593235 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 9 |
Hb_001673_110 |
0.2290923232 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 10 |
Hb_022593_030 |
0.232792889 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 11 |
Hb_000035_300 |
0.2394642043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005147_050 |
0.2429833902 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 13 |
Hb_002609_150 |
0.2436265587 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006615_040 |
0.2437079475 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 15 |
Hb_151281_010 |
0.2490345278 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 16 |
Hb_138628_010 |
0.250485342 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_012018_010 |
0.2506620214 |
- |
- |
hypothetical protein MIMGU_mgv1a0024251mg, partial [Erythranthe guttata] |
| 18 |
Hb_079955_010 |
0.2523892325 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 19 |
Hb_001584_010 |
0.2533959395 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 20 |
Hb_116635_010 |
0.2538063435 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |