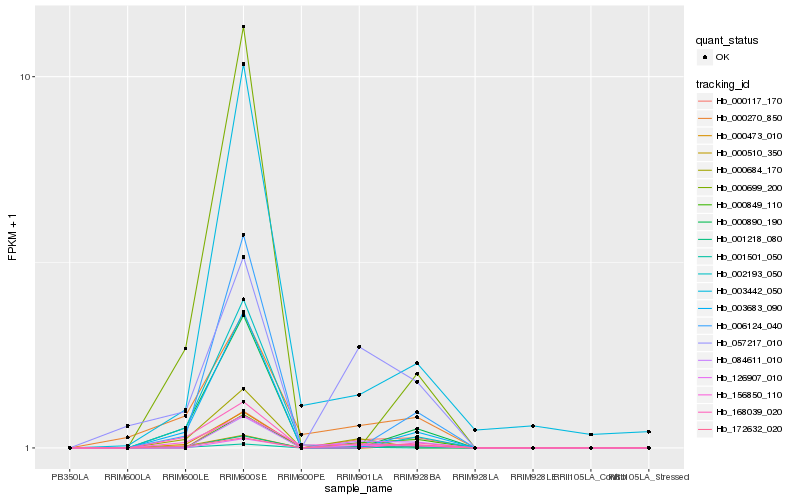
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001218_080 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 2 |
Hb_156850_110 |
0.1788723826 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 3 |
Hb_000684_170 |
0.1917362446 |
- |
- |
- |
| 4 |
Hb_001501_050 |
0.1974173242 |
- |
- |
Wall associated kinase-like 6, putative [Theobroma cacao] |
| 5 |
Hb_172632_020 |
0.2120882659 |
- |
- |
- |
| 6 |
Hb_000270_850 |
0.2153115412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_084611_010 |
0.2196591499 |
- |
- |
PREDICTED: uncharacterized protein LOC104249812 [Nicotiana sylvestris] |
| 8 |
Hb_000890_190 |
0.223152656 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 9 |
Hb_003683_090 |
0.2258412152 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_057217_010 |
0.2267495041 |
- |
- |
PREDICTED: uncharacterized protein LOC104594412 [Nelumbo nucifera] |
| 11 |
Hb_000510_350 |
0.2325859565 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 12 |
Hb_000849_110 |
0.2342316383 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 13 |
Hb_000699_200 |
0.2366414979 |
- |
- |
hypothetical protein POPTR_0019s02340g [Populus trichocarpa] |
| 14 |
Hb_000473_010 |
0.2369043784 |
- |
- |
PREDICTED: galactolipase DONGLE, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003442_050 |
0.2391512859 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 16 |
Hb_168039_020 |
0.241550317 |
- |
- |
- |
| 17 |
Hb_126907_010 |
0.2434863092 |
- |
- |
- |
| 18 |
Hb_002193_050 |
0.2439830994 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 19 |
Hb_000117_170 |
0.2445459805 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 20 |
Hb_006124_040 |
0.2504816958 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |