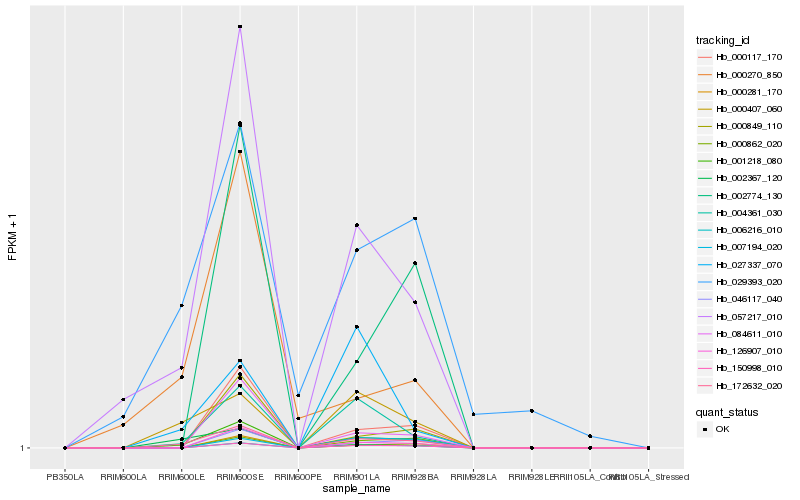
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172632_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002367_120 |
0.1395239696 |
- |
- |
- |
| 3 |
Hb_057217_010 |
0.1467346898 |
- |
- |
PREDICTED: uncharacterized protein LOC104594412 [Nelumbo nucifera] |
| 4 |
Hb_000407_060 |
0.1852662681 |
- |
- |
- |
| 5 |
Hb_004361_030 |
0.1951687642 |
- |
- |
PREDICTED: uncharacterized protein LOC104239397 isoform X1 [Nicotiana sylvestris] |
| 6 |
Hb_001218_080 |
0.2120882659 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 7 |
Hb_150998_010 |
0.2190693289 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 8 |
Hb_006216_010 |
0.2217931843 |
- |
- |
PREDICTED: uncharacterized protein LOC103411650 [Malus domestica] |
| 9 |
Hb_126907_010 |
0.2246242391 |
- |
- |
- |
| 10 |
Hb_046117_040 |
0.2289043512 |
- |
- |
hypothetical protein JCGZ_07941 [Jatropha curcas] |
| 11 |
Hb_000117_170 |
0.2353012088 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 12 |
Hb_000281_170 |
0.236760838 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_084611_010 |
0.2461966551 |
- |
- |
PREDICTED: uncharacterized protein LOC104249812 [Nicotiana sylvestris] |
| 14 |
Hb_002774_130 |
0.2595260705 |
- |
- |
- |
| 15 |
Hb_000849_110 |
0.2611991206 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 16 |
Hb_000270_850 |
0.2722212326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000862_020 |
0.2734213324 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 18 |
Hb_027337_070 |
0.276356484 |
- |
- |
hypothetical protein POPTR_0001s29030g [Populus trichocarpa] |
| 19 |
Hb_029393_020 |
0.2790688031 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 20 |
Hb_007194_020 |
0.2922177376 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |