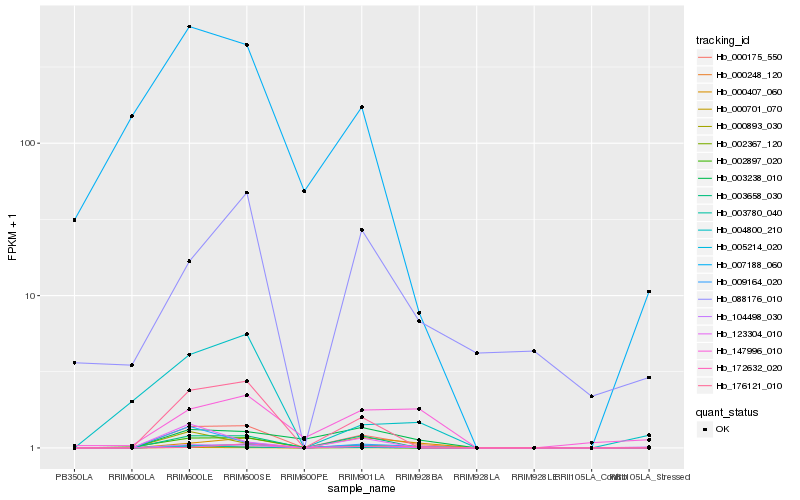
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_550 |
0.0 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 2 |
Hb_176121_010 |
0.1732455603 |
- |
- |
- |
| 3 |
Hb_003658_030 |
0.1931687226 |
- |
- |
- |
| 4 |
Hb_000701_070 |
0.2051145578 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_002367_120 |
0.2258659987 |
- |
- |
- |
| 6 |
Hb_005214_020 |
0.231509389 |
- |
- |
hypothetical protein JCGZ_12886 [Jatropha curcas] |
| 7 |
Hb_104498_030 |
0.2434565933 |
- |
- |
hypothetical protein JCGZ_11092 [Jatropha curcas] |
| 8 |
Hb_000248_120 |
0.2615253131 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 9 |
Hb_000407_060 |
0.2615966577 |
- |
- |
- |
| 10 |
Hb_009164_020 |
0.2938085461 |
- |
- |
hypothetical protein PRUPE_ppa026212mg, partial [Prunus persica] |
| 11 |
Hb_003238_010 |
0.3003329726 |
- |
- |
PREDICTED: uncharacterized protein LOC104880636 [Vitis vinifera] |
| 12 |
Hb_123304_010 |
0.3086991033 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 13 |
Hb_003780_040 |
0.3155118185 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 14 |
Hb_172632_020 |
0.316433479 |
- |
- |
- |
| 15 |
Hb_000893_030 |
0.31802513 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 16 |
Hb_088176_010 |
0.3235719664 |
- |
- |
- |
| 17 |
Hb_004800_210 |
0.3299772402 |
- |
- |
hypothetical protein POPTR_0007s12780g [Populus trichocarpa] |
| 18 |
Hb_007188_060 |
0.3310720148 |
- |
- |
- |
| 19 |
Hb_147996_010 |
0.3353456919 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002897_020 |
0.3412143397 |
- |
- |
phosphate transporter [Manihot esculenta] |