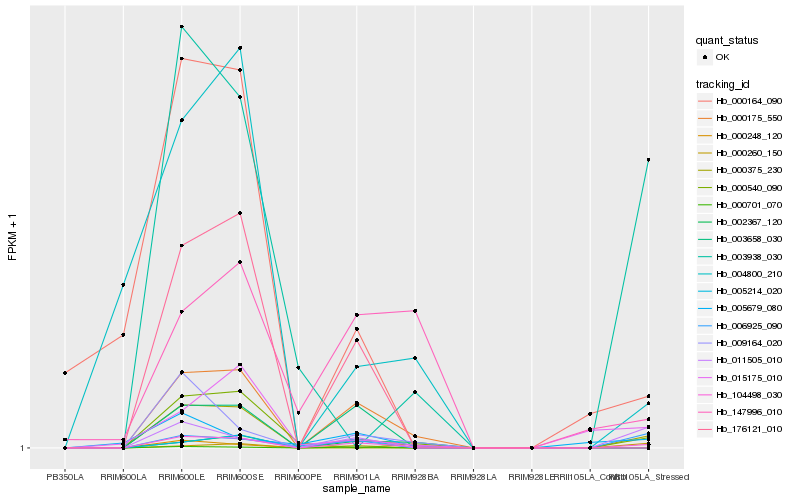
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104498_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_11092 [Jatropha curcas] |
| 2 |
Hb_000175_550 |
0.2434565933 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 3 |
Hb_011505_010 |
0.2849594419 |
- |
- |
- |
| 4 |
Hb_176121_010 |
0.2963085588 |
- |
- |
- |
| 5 |
Hb_000701_070 |
0.3073648876 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_006925_090 |
0.3121486623 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 7 |
Hb_003658_030 |
0.3157002512 |
- |
- |
- |
| 8 |
Hb_000260_150 |
0.321710058 |
- |
- |
hypothetical protein POPTR_0344s002001g, partial [Populus trichocarpa] |
| 9 |
Hb_000375_230 |
0.3343777821 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 10 |
Hb_000540_090 |
0.3344192813 |
- |
- |
PREDICTED: chitinase 2-like [Jatropha curcas] |
| 11 |
Hb_003938_030 |
0.3368947214 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 12 |
Hb_004800_210 |
0.3382027906 |
- |
- |
hypothetical protein POPTR_0007s12780g [Populus trichocarpa] |
| 13 |
Hb_147996_010 |
0.3389651344 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009164_020 |
0.3403871067 |
- |
- |
hypothetical protein PRUPE_ppa026212mg, partial [Prunus persica] |
| 15 |
Hb_000164_090 |
0.3440074055 |
- |
- |
hypothetical protein [Plasmodium berghei strain ANKA], partial [Plasmodium berghei ANKA] |
| 16 |
Hb_002367_120 |
0.3440682628 |
- |
- |
- |
| 17 |
Hb_000248_120 |
0.3520966208 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 18 |
Hb_005679_080 |
0.3532533946 |
- |
- |
PREDICTED: PH-interacting protein-like [Populus euphratica] |
| 19 |
Hb_005214_020 |
0.3539928866 |
- |
- |
hypothetical protein JCGZ_12886 [Jatropha curcas] |
| 20 |
Hb_015175_010 |
0.3615173665 |
- |
- |
hypothetical protein JCGZ_17977 [Jatropha curcas] |