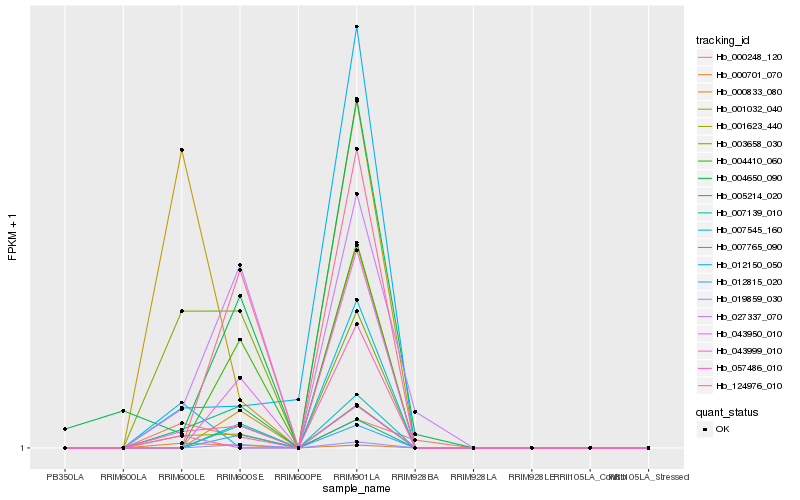
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_440 |
0.0 |
- |
- |
hypothetical protein JCGZ_12908 [Jatropha curcas] |
| 2 |
Hb_057486_010 |
0.1411225431 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 3 |
Hb_003658_030 |
0.2508535891 |
- |
- |
- |
| 4 |
Hb_027337_070 |
0.2643134395 |
- |
- |
hypothetical protein POPTR_0001s29030g [Populus trichocarpa] |
| 5 |
Hb_001032_040 |
0.2674794303 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005214_020 |
0.2827031956 |
- |
- |
hypothetical protein JCGZ_12886 [Jatropha curcas] |
| 7 |
Hb_012150_050 |
0.3063861822 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_000248_120 |
0.3072058786 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 9 |
Hb_043950_010 |
0.3080500001 |
- |
- |
PREDICTED: uncharacterized protein LOC102619199, partial [Citrus sinensis] |
| 10 |
Hb_007545_160 |
0.311616232 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 11 |
Hb_000701_070 |
0.3123042767 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_004410_060 |
0.318526897 |
- |
- |
- |
| 13 |
Hb_004650_090 |
0.3222671157 |
- |
- |
- |
| 14 |
Hb_124976_010 |
0.3239149427 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 15 |
Hb_000833_080 |
0.3244677811 |
- |
- |
PREDICTED: uncharacterized protein LOC105789525 [Gossypium raimondii] |
| 16 |
Hb_019859_030 |
0.3260479289 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 17 |
Hb_007139_010 |
0.3261372015 |
- |
- |
hypothetical protein POPTR_0016s134801g, partial [Populus trichocarpa] |
| 18 |
Hb_043999_010 |
0.326263911 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 19 |
Hb_007765_090 |
0.3262686099 |
- |
- |
unknown [Glycine max] |
| 20 |
Hb_012815_020 |
0.3263642254 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |