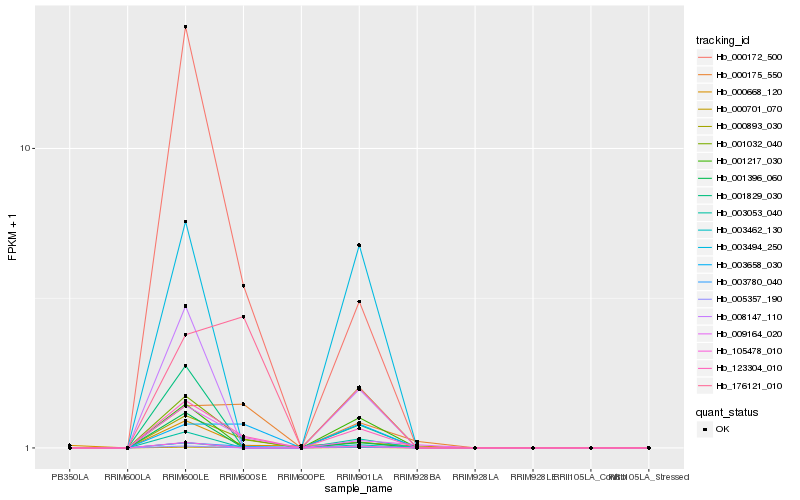
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123304_010 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 2 |
Hb_000893_030 |
0.1322428397 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 3 |
Hb_000701_070 |
0.1586043786 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_009164_020 |
0.2018588024 |
- |
- |
hypothetical protein PRUPE_ppa026212mg, partial [Prunus persica] |
| 5 |
Hb_000172_500 |
0.217341963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001032_040 |
0.2566595116 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003658_030 |
0.2675414522 |
- |
- |
- |
| 8 |
Hb_105478_010 |
0.2681363792 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 9 |
Hb_008147_110 |
0.2706146746 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_003053_040 |
0.2769491378 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001829_030 |
0.2810600305 |
- |
- |
PREDICTED: uncharacterized protein LOC105119087 [Populus euphratica] |
| 12 |
Hb_003462_130 |
0.2817221237 |
- |
- |
- |
| 13 |
Hb_005357_190 |
0.2885622423 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 14 |
Hb_001217_030 |
0.2888745793 |
- |
- |
- |
| 15 |
Hb_003780_040 |
0.306218598 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 16 |
Hb_003494_250 |
0.3064721953 |
- |
- |
- |
| 17 |
Hb_000175_550 |
0.3086991033 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 18 |
Hb_001396_060 |
0.3097230442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000668_120 |
0.3130397678 |
- |
- |
hypothetical protein JCGZ_11991 [Jatropha curcas] |
| 20 |
Hb_176121_010 |
0.3155250844 |
- |
- |
- |