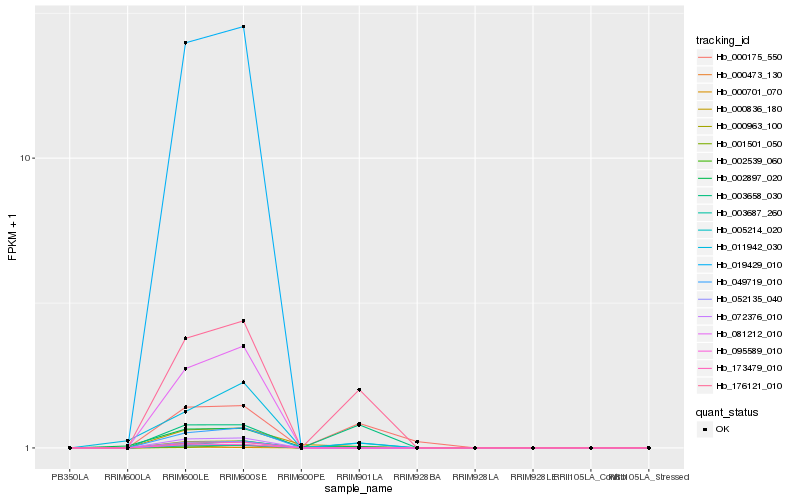
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176121_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000175_550 |
0.1732455603 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 3 |
Hb_003658_030 |
0.1787419889 |
- |
- |
- |
| 4 |
Hb_005214_020 |
0.1941624727 |
- |
- |
hypothetical protein JCGZ_12886 [Jatropha curcas] |
| 5 |
Hb_000701_070 |
0.1976673847 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_011942_030 |
0.2556274095 |
- |
- |
- |
| 7 |
Hb_002897_020 |
0.2656165849 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_000836_180 |
0.2686828618 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 9 |
Hb_001501_050 |
0.285605599 |
- |
- |
Wall associated kinase-like 6, putative [Theobroma cacao] |
| 10 |
Hb_173479_010 |
0.2900174585 |
- |
- |
- |
| 11 |
Hb_003687_260 |
0.2902908819 |
- |
- |
PREDICTED: inorganic phosphate transporter 1-4-like [Camelina sativa] |
| 12 |
Hb_002539_060 |
0.2903325499 |
- |
- |
- |
| 13 |
Hb_049719_010 |
0.2905386699 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_019429_010 |
0.2906550968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_072376_010 |
0.2910136881 |
- |
- |
- |
| 16 |
Hb_000963_100 |
0.2910359452 |
- |
- |
Protein SUA5, putative [Ricinus communis] |
| 17 |
Hb_052135_040 |
0.2910408804 |
- |
- |
- |
| 18 |
Hb_081212_010 |
0.2910411417 |
- |
- |
- |
| 19 |
Hb_095589_010 |
0.2910624725 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 20 |
Hb_000473_130 |
0.2910725974 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |