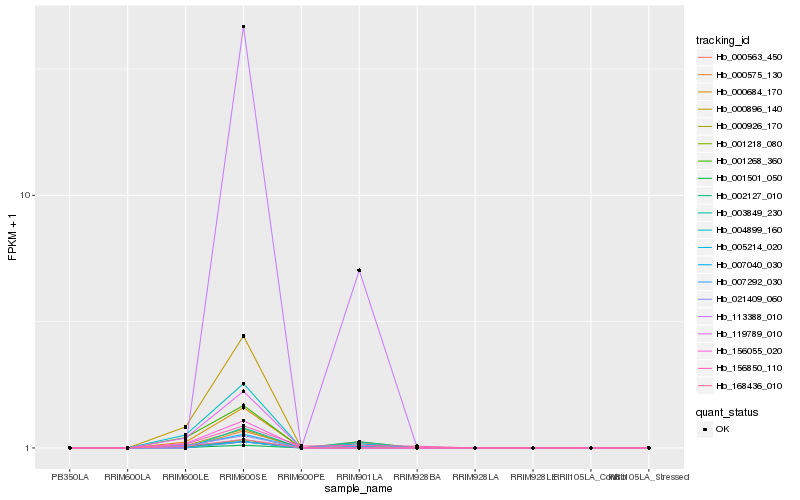
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001501_050 |
0.0 |
- |
- |
Wall associated kinase-like 6, putative [Theobroma cacao] |
| 2 |
Hb_000684_170 |
0.035267203 |
- |
- |
- |
| 3 |
Hb_001218_080 |
0.1974173242 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 4 |
Hb_156850_110 |
0.2408271137 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 5 |
Hb_007292_030 |
0.2424138786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_113388_010 |
0.2522155222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003849_230 |
0.2533747653 |
- |
- |
PREDICTED: NEP1-interacting protein 2-like [Jatropha curcas] |
| 8 |
Hb_000575_130 |
0.2536043901 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Populus euphratica] |
| 9 |
Hb_000563_450 |
0.2544074809 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 10 |
Hb_005214_020 |
0.2559287354 |
- |
- |
hypothetical protein JCGZ_12886 [Jatropha curcas] |
| 11 |
Hb_119789_010 |
0.2591144253 |
- |
- |
PREDICTED: uncharacterized protein LOC105134438 [Populus euphratica] |
| 12 |
Hb_156055_020 |
0.2591515957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004899_160 |
0.2594154773 |
- |
- |
- |
| 14 |
Hb_000896_140 |
0.2602135756 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_007040_030 |
0.2602691289 |
- |
- |
PREDICTED: uncharacterized protein LOC103415345 [Malus domestica] |
| 16 |
Hb_000926_170 |
0.2609479268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_021409_060 |
0.2611969989 |
- |
- |
PREDICTED: aluminum-activated malate transporter 10-like [Jatropha curcas] |
| 18 |
Hb_168436_010 |
0.2618113757 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000-like [Citrus sinensis] |
| 19 |
Hb_002127_010 |
0.2631182216 |
- |
- |
Aluminum activated malate transporter family protein, putative [Theobroma cacao] |
| 20 |
Hb_001268_360 |
0.264791925 |
- |
- |
PREDICTED: uncharacterized protein LOC104447679 [Eucalyptus grandis] |