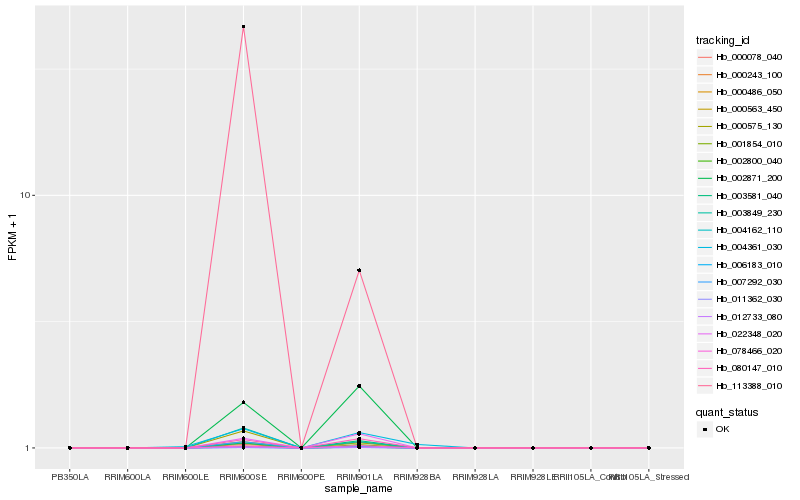
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000486_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_000563_450 |
0.076562965 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 3 |
Hb_000575_130 |
0.0789908199 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Populus euphratica] |
| 4 |
Hb_003849_230 |
0.0796955853 |
- |
- |
PREDICTED: NEP1-interacting protein 2-like [Jatropha curcas] |
| 5 |
Hb_001854_010 |
0.0913094 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_007292_030 |
0.1234509374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000243_100 |
0.1417384849 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 8 |
Hb_004162_110 |
0.1417681602 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 9 |
Hb_002800_040 |
0.142146413 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_011362_030 |
0.1810017813 |
- |
- |
polyprotein [Oryza australiensis] |
| 11 |
Hb_012733_080 |
0.1893447029 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_006183_010 |
0.1901367384 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_080147_010 |
0.1912645499 |
- |
- |
- |
| 14 |
Hb_078466_020 |
0.2230988985 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 15 |
Hb_113388_010 |
0.2476877861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002871_200 |
0.2489490023 |
- |
- |
- |
| 17 |
Hb_003581_040 |
0.2499945279 |
- |
- |
hypothetical protein PHAVU_002G080900g [Phaseolus vulgaris] |
| 18 |
Hb_022348_020 |
0.2520573638 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_004361_030 |
0.2693930923 |
- |
- |
PREDICTED: uncharacterized protein LOC104239397 isoform X1 [Nicotiana sylvestris] |
| 20 |
Hb_000078_040 |
0.2709835022 |
- |
- |
PREDICTED: uncharacterized protein LOC104907366 [Beta vulgaris subsp. vulgaris] |