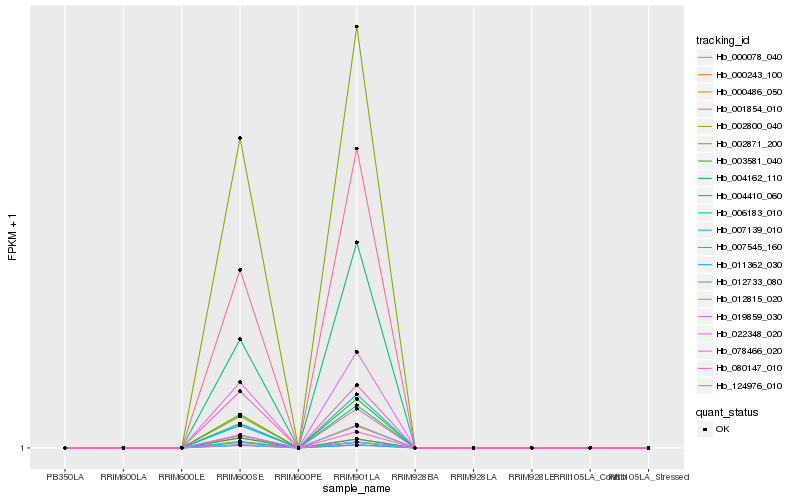
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 2 |
Hb_000243_100 |
2.98966e-05 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 3 |
Hb_002800_040 |
0.0003810801 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_011362_030 |
0.0396006498 |
- |
- |
polyprotein [Oryza australiensis] |
| 5 |
Hb_012733_080 |
0.048040631 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_006183_010 |
0.0488422249 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_080147_010 |
0.0499837498 |
- |
- |
- |
| 8 |
Hb_001854_010 |
0.0507078953 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 9 |
Hb_078466_020 |
0.0822549796 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 10 |
Hb_002871_200 |
0.1085308281 |
- |
- |
- |
| 11 |
Hb_003581_040 |
0.1095949225 |
- |
- |
hypothetical protein PHAVU_002G080900g [Phaseolus vulgaris] |
| 12 |
Hb_022348_020 |
0.1116947047 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 13 |
Hb_000078_040 |
0.1309791193 |
- |
- |
PREDICTED: uncharacterized protein LOC104907366 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000486_050 |
0.1417681602 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_012815_020 |
0.144891605 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 16 |
Hb_007139_010 |
0.1456169237 |
- |
- |
hypothetical protein POPTR_0016s134801g, partial [Populus trichocarpa] |
| 17 |
Hb_019859_030 |
0.1459032492 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 18 |
Hb_124976_010 |
0.1529444884 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 19 |
Hb_004410_060 |
0.1729595994 |
- |
- |
- |
| 20 |
Hb_007545_160 |
0.2082699255 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |