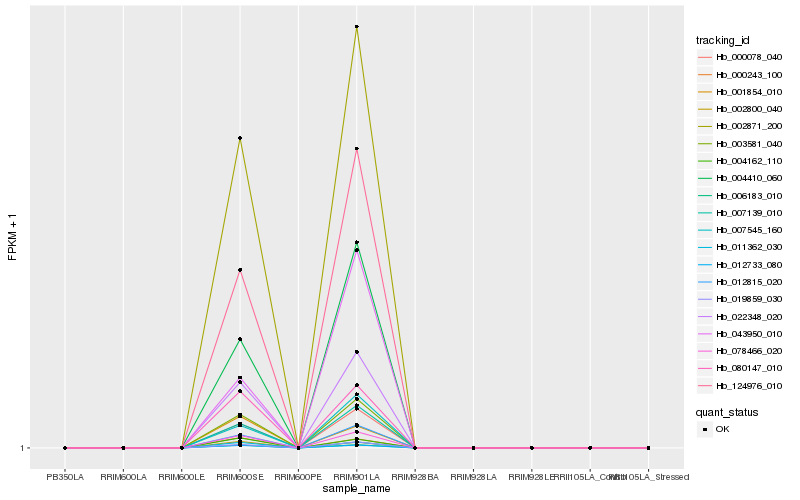
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078466_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 2 |
Hb_002871_200 |
0.0263586455 |
- |
- |
- |
| 3 |
Hb_003581_040 |
0.0274269659 |
- |
- |
hypothetical protein PHAVU_002G080900g [Phaseolus vulgaris] |
| 4 |
Hb_022348_020 |
0.0295352891 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 5 |
Hb_080147_010 |
0.0323174354 |
- |
- |
- |
| 6 |
Hb_006183_010 |
0.0334594939 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_012733_080 |
0.0342614179 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_011362_030 |
0.0427026433 |
- |
- |
polyprotein [Oryza australiensis] |
| 9 |
Hb_000078_040 |
0.0489108155 |
- |
- |
PREDICTED: uncharacterized protein LOC104907366 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_012815_020 |
0.062903492 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 11 |
Hb_007139_010 |
0.0636333303 |
- |
- |
hypothetical protein POPTR_0016s134801g, partial [Populus trichocarpa] |
| 12 |
Hb_019859_030 |
0.0639214492 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 13 |
Hb_124976_010 |
0.0710084624 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_002800_040 |
0.0818747891 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_004162_110 |
0.0822549796 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 16 |
Hb_000243_100 |
0.0822848061 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 17 |
Hb_004410_060 |
0.0911715327 |
- |
- |
- |
| 18 |
Hb_007545_160 |
0.1268101991 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 19 |
Hb_001854_010 |
0.1327703284 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 20 |
Hb_043950_010 |
0.1703645515 |
- |
- |
PREDICTED: uncharacterized protein LOC102619199, partial [Citrus sinensis] |