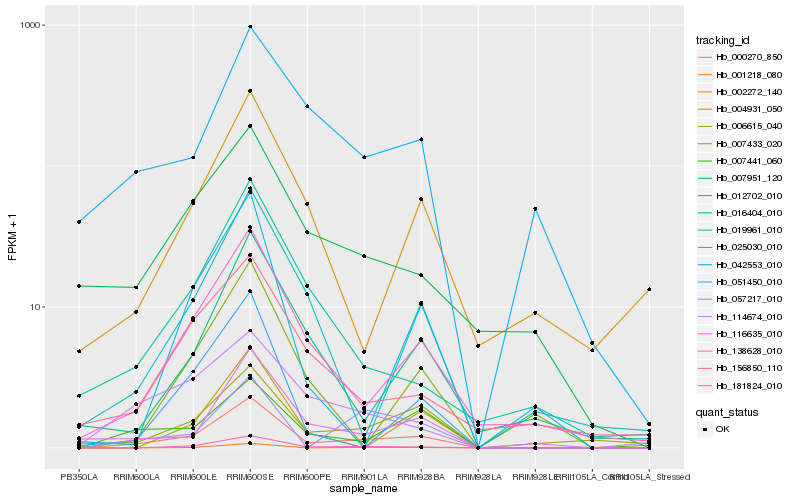
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_850 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_156850_110 |
0.1583708571 |
- |
- |
PREDICTED: vinorine synthase-like [Jatropha curcas] |
| 3 |
Hb_138628_010 |
0.1895126043 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 4 |
Hb_007441_060 |
0.1953716059 |
- |
- |
- |
| 5 |
Hb_116635_010 |
0.2080995077 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_001218_080 |
0.2153115412 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 7 |
Hb_006615_040 |
0.2190430255 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 8 |
Hb_051450_010 |
0.2331483645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_057217_010 |
0.2339942092 |
- |
- |
PREDICTED: uncharacterized protein LOC104594412 [Nelumbo nucifera] |
| 10 |
Hb_114674_010 |
0.2348526368 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_019961_010 |
0.2354618339 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 12 |
Hb_042553_010 |
0.2388185246 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 13 |
Hb_012702_010 |
0.2390272999 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 14 |
Hb_007951_120 |
0.2391398512 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 15 |
Hb_004931_050 |
0.2423949888 |
transcription factor |
TF Family: MYB-related |
Transcription repressor MYB4, putative [Ricinus communis] |
| 16 |
Hb_181824_010 |
0.2449309227 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_007433_020 |
0.2450003485 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 18 |
Hb_016404_010 |
0.2508121112 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 19 |
Hb_002272_140 |
0.2515773534 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 20 |
Hb_025030_010 |
0.2523431611 |
- |
- |
- |