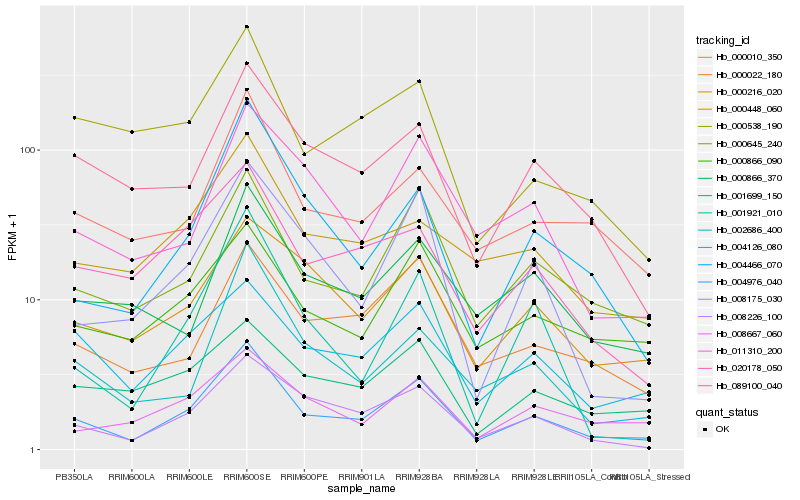
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004976_040 |
0.0 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 2 |
Hb_000645_240 |
0.1268053748 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 3 |
Hb_008226_100 |
0.1315701826 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 4 |
Hb_000010_350 |
0.1494642517 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 5 |
Hb_000866_090 |
0.150619883 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_004126_080 |
0.1519703344 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 7 |
Hb_089100_040 |
0.1534473967 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 8 |
Hb_000216_020 |
0.1554322937 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 9 |
Hb_008175_030 |
0.1555812176 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 10 |
Hb_000022_180 |
0.1562463821 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_020178_050 |
0.1588868782 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 12 |
Hb_004466_070 |
0.1598264958 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 13 |
Hb_000448_060 |
0.1630794586 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 14 |
Hb_008667_060 |
0.1634776714 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 15 |
Hb_011310_200 |
0.1641501267 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 16 |
Hb_002686_400 |
0.1651099406 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 17 |
Hb_001699_150 |
0.1674036491 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 18 |
Hb_001921_010 |
0.1675608918 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000866_370 |
0.1683757087 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 20 |
Hb_000538_190 |
0.1688183474 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |