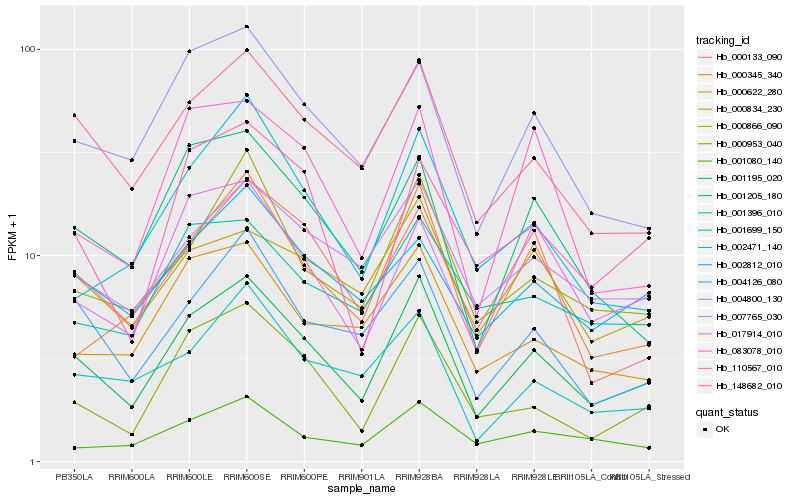
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_020 |
0.0 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_148682_010 |
0.1022989323 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 3 |
Hb_000953_040 |
0.1146308201 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 4 |
Hb_000866_090 |
0.1152401391 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_007765_030 |
0.1158297967 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 6 |
Hb_004800_130 |
0.1181162983 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 7 |
Hb_004126_080 |
0.1189658292 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 8 |
Hb_001205_180 |
0.1266048405 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 9 |
Hb_000834_230 |
0.128306599 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 10 |
Hb_000622_280 |
0.1303812181 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001699_150 |
0.130442866 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 12 |
Hb_110567_010 |
0.1307682133 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000345_340 |
0.1314178414 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 14 |
Hb_000133_090 |
0.1328770656 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 15 |
Hb_002812_010 |
0.1335092333 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_017914_010 |
0.1337026547 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 17 |
Hb_001080_140 |
0.134417719 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001396_010 |
0.1386043018 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_002471_140 |
0.1386190854 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 20 |
Hb_083078_010 |
0.1399520349 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |