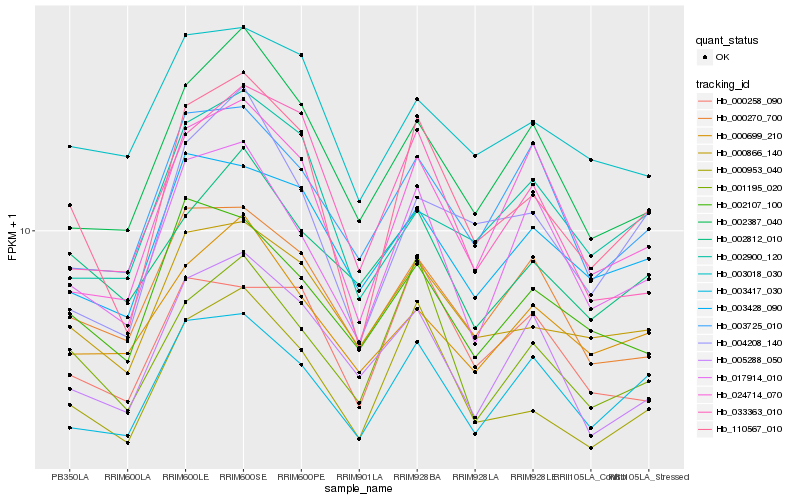
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110567_010 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000953_040 |
0.1098577069 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 3 |
Hb_000866_140 |
0.1191240586 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 4 |
Hb_003417_030 |
0.1269015133 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001195_020 |
0.1307682133 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_024714_070 |
0.1329480862 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 7 |
Hb_017914_010 |
0.1333041946 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 8 |
Hb_004208_140 |
0.1359765934 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 9 |
Hb_002387_040 |
0.1369957171 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_033363_010 |
0.1371115038 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 11 |
Hb_003428_090 |
0.143626987 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 12 |
Hb_000270_700 |
0.1450208826 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 13 |
Hb_002107_100 |
0.1491169567 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_000258_090 |
0.1499031776 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002900_120 |
0.1507410883 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 16 |
Hb_003018_030 |
0.1510898417 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 17 |
Hb_003725_010 |
0.1541899826 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 18 |
Hb_005288_050 |
0.1553007969 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 19 |
Hb_002812_010 |
0.1555040887 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000699_210 |
0.155913817 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |