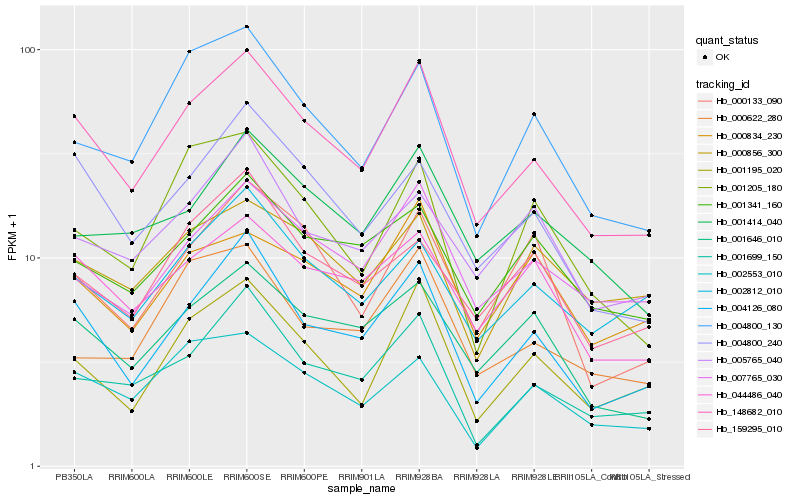
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148682_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 2 |
Hb_004126_080 |
0.0744084493 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 3 |
Hb_001646_010 |
0.0988008377 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 4 |
Hb_004800_240 |
0.0989726371 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 5 |
Hb_001195_020 |
0.1022989323 |
- |
- |
PREDICTED: ornithine carbamoyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001699_150 |
0.1026557373 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 7 |
Hb_044486_040 |
0.1027739735 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 8 |
Hb_007765_030 |
0.1044697901 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 9 |
Hb_004800_130 |
0.1081584546 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 10 |
Hb_001414_040 |
0.1127796409 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001341_160 |
0.1135876443 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002553_010 |
0.114121698 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 13 |
Hb_002812_010 |
0.117222864 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000622_280 |
0.1216769272 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000834_230 |
0.1224305484 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 16 |
Hb_000133_090 |
0.1227462122 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 17 |
Hb_005765_040 |
0.1245345578 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 18 |
Hb_159295_010 |
0.1252976132 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 19 |
Hb_001205_180 |
0.1263090127 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 20 |
Hb_000856_300 |
0.1264489626 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |