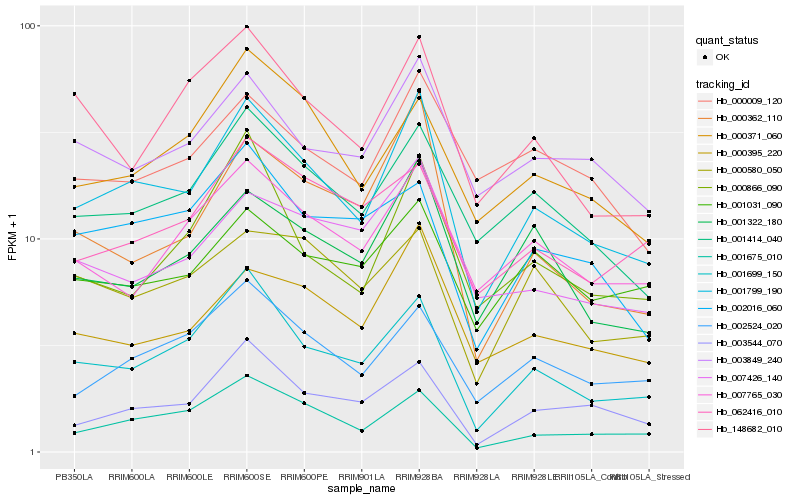
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001799_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 2 |
Hb_001699_150 |
0.0986663563 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 3 |
Hb_001414_040 |
0.107619504 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001675_010 |
0.1084169104 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 5 |
Hb_000395_220 |
0.1129318802 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 6 |
Hb_001322_180 |
0.1161695177 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 7 |
Hb_002524_020 |
0.1185762559 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 8 |
Hb_003544_070 |
0.1190118312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003849_240 |
0.122204461 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_007765_030 |
0.1263525314 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 11 |
Hb_000362_110 |
0.1265370671 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 12 |
Hb_000371_060 |
0.128833483 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 13 |
Hb_000866_090 |
0.1298796618 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_148682_010 |
0.1312750825 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 15 |
Hb_002016_060 |
0.1321357263 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 16 |
Hb_007426_140 |
0.1324514045 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 17 |
Hb_001031_090 |
0.137588641 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 18 |
Hb_000009_120 |
0.1380306685 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 19 |
Hb_000580_050 |
0.1399135135 |
- |
- |
- |
| 20 |
Hb_062416_010 |
0.1407157328 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |