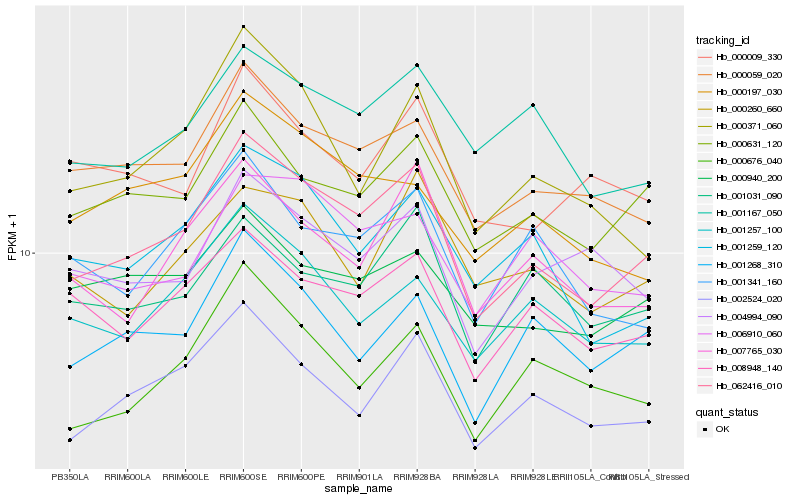
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062416_010 |
0.0 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 2 |
Hb_000631_120 |
0.0898814006 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 3 |
Hb_000059_020 |
0.0916993424 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 4 |
Hb_002524_020 |
0.0980604045 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 5 |
Hb_008948_140 |
0.1007952316 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 6 |
Hb_007765_030 |
0.1024508811 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 7 |
Hb_001259_120 |
0.1028321336 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 8 |
Hb_001167_050 |
0.1060161511 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 9 |
Hb_001268_310 |
0.106939822 |
- |
- |
- |
| 10 |
Hb_001257_100 |
0.1084570174 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 11 |
Hb_000940_200 |
0.1091445592 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 12 |
Hb_001031_090 |
0.1099248401 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 13 |
Hb_006910_060 |
0.1108916805 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 14 |
Hb_000371_060 |
0.1125464889 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 15 |
Hb_000260_660 |
0.1130409316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004994_090 |
0.1134834239 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001341_160 |
0.1135051948 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000009_330 |
0.1155025166 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_000676_040 |
0.1160610746 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 20 |
Hb_000197_030 |
0.1162255046 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |