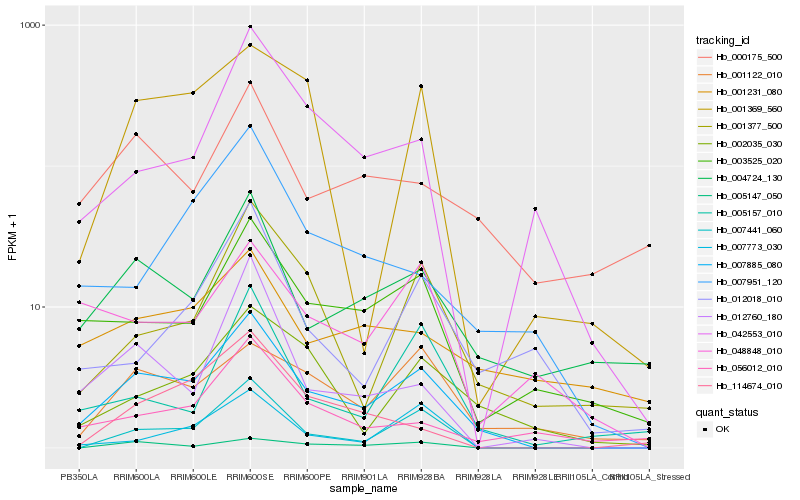
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007885_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007441_060 |
0.1904291533 |
- |
- |
- |
| 3 |
Hb_004724_130 |
0.1987128522 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 4 |
Hb_003525_020 |
0.2029471656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002035_030 |
0.2248775986 |
- |
- |
hypothetical protein PRUPE_ppa003221mg [Prunus persica] |
| 6 |
Hb_005147_050 |
0.2256762463 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 7 |
Hb_001122_010 |
0.2298813428 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 8 |
Hb_001377_500 |
0.2327538196 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 9 |
Hb_007773_030 |
0.2337133676 |
- |
- |
- |
| 10 |
Hb_000175_500 |
0.2339911801 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 11 |
Hb_012760_180 |
0.239794653 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 12 |
Hb_042553_010 |
0.2401067156 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 13 |
Hb_007951_120 |
0.2402929772 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 14 |
Hb_056012_010 |
0.2413134784 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 15 |
Hb_012018_010 |
0.2436134616 |
- |
- |
hypothetical protein MIMGU_mgv1a0024251mg, partial [Erythranthe guttata] |
| 16 |
Hb_005157_010 |
0.2443364625 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 17 |
Hb_001369_560 |
0.2444154367 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 18 |
Hb_001231_080 |
0.2455919884 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_048848_010 |
0.2457197439 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 20 |
Hb_114674_010 |
0.2483510539 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |