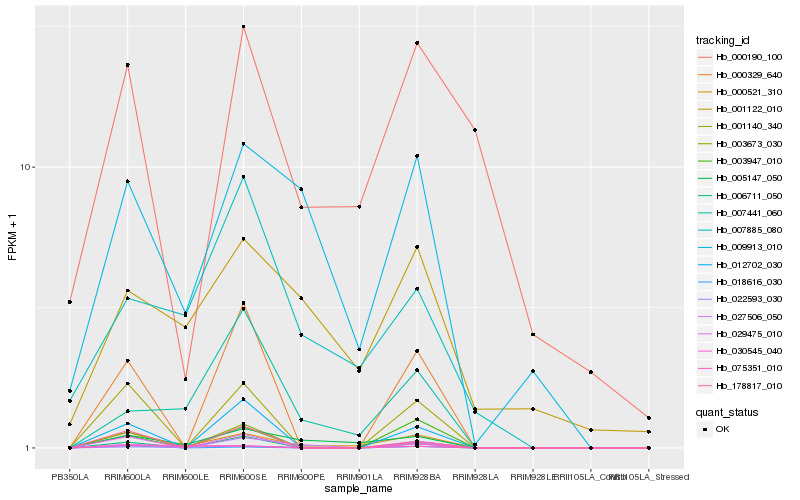
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_310 |
0.0 |
- |
- |
- |
| 2 |
Hb_005147_050 |
0.1936740929 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 3 |
Hb_001140_340 |
0.2493255518 |
- |
- |
- |
| 4 |
Hb_009913_010 |
0.259645189 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_029475_010 |
0.2637308876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_022593_030 |
0.2758822318 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_018616_030 |
0.2808144401 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 8 |
Hb_000329_640 |
0.2863648115 |
- |
- |
- |
| 9 |
Hb_007441_060 |
0.2964746534 |
- |
- |
- |
| 10 |
Hb_006711_050 |
0.2978622414 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 11 |
Hb_012702_030 |
0.2999280128 |
- |
- |
hypothetical protein POPTR_0077s002001g, partial [Populus trichocarpa] |
| 12 |
Hb_030545_040 |
0.3001325211 |
- |
- |
- |
| 13 |
Hb_003947_010 |
0.3031690724 |
- |
- |
hypothetical protein JCGZ_24829 [Jatropha curcas] |
| 14 |
Hb_001122_010 |
0.3032332478 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 15 |
Hb_178817_010 |
0.3064716528 |
- |
- |
protease [Gossypium hirsutum] |
| 16 |
Hb_007885_080 |
0.3071283046 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 17 |
Hb_027506_050 |
0.3132662007 |
- |
- |
- |
| 18 |
Hb_003673_030 |
0.3133791259 |
- |
- |
hypothetical protein POPTR_0011s11940g [Populus trichocarpa] |
| 19 |
Hb_000190_100 |
0.3168153049 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 20 |
Hb_075351_010 |
0.320006142 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |