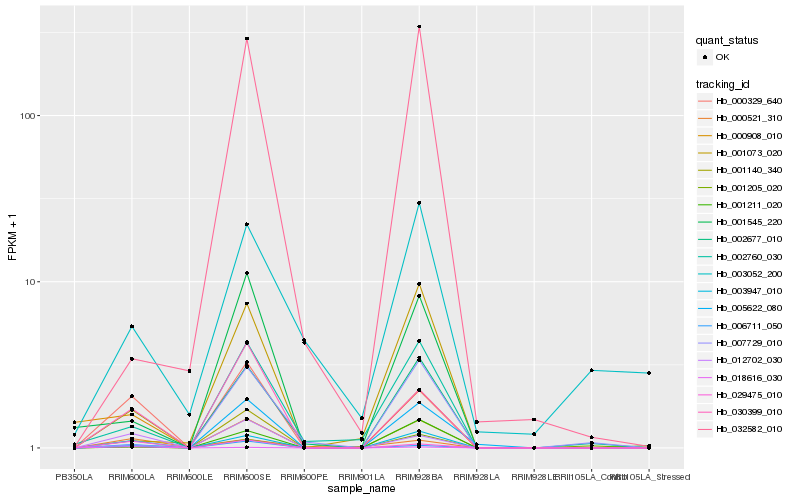
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003947_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_24829 [Jatropha curcas] |
| 2 |
Hb_000329_640 |
0.1820669647 |
- |
- |
- |
| 3 |
Hb_001140_340 |
0.2143576929 |
- |
- |
- |
| 4 |
Hb_012702_030 |
0.2383257498 |
- |
- |
hypothetical protein POPTR_0077s002001g, partial [Populus trichocarpa] |
| 5 |
Hb_029475_010 |
0.24457243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002760_030 |
0.2479092685 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 7 |
Hb_006711_050 |
0.249265954 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 8 |
Hb_030399_010 |
0.2497624445 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 9 |
Hb_018616_030 |
0.259359067 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 10 |
Hb_001211_020 |
0.2834343037 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 11 |
Hb_001545_220 |
0.2891538431 |
- |
- |
hypothetical protein POPTR_0002s06170g [Populus trichocarpa] |
| 12 |
Hb_001205_020 |
0.2902133003 |
- |
- |
- |
| 13 |
Hb_005622_080 |
0.2954270094 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 14 |
Hb_001073_020 |
0.2997512304 |
- |
- |
hypothetical protein CISIN_1g031854mg [Citrus sinensis] |
| 15 |
Hb_003052_200 |
0.3030563981 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 16 |
Hb_000521_310 |
0.3031690724 |
- |
- |
- |
| 17 |
Hb_002677_010 |
0.3049630416 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 18 |
Hb_000908_010 |
0.3091136024 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 19 |
Hb_032582_010 |
0.3092545681 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 20 |
Hb_007729_010 |
0.3104362178 |
- |
- |
unnamed protein product [Vitis vinifera] |