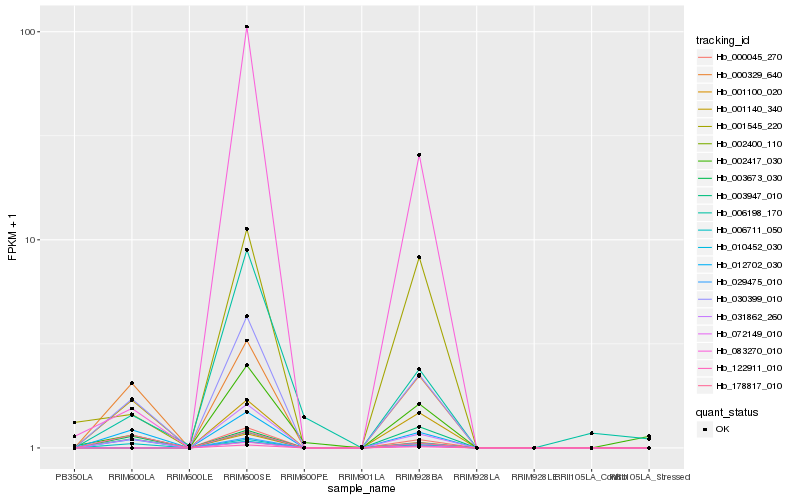
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030399_010 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 2 |
Hb_031862_260 |
0.0656484227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012702_030 |
0.1176007616 |
- |
- |
hypothetical protein POPTR_0077s002001g, partial [Populus trichocarpa] |
| 4 |
Hb_000329_640 |
0.1189611802 |
- |
- |
- |
| 5 |
Hb_006711_050 |
0.1342535192 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 6 |
Hb_003673_030 |
0.2133312662 |
- |
- |
hypothetical protein POPTR_0011s11940g [Populus trichocarpa] |
| 7 |
Hb_029475_010 |
0.2241004395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002417_030 |
0.2362361877 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 9 |
Hb_001545_220 |
0.2364874644 |
- |
- |
hypothetical protein POPTR_0002s06170g [Populus trichocarpa] |
| 10 |
Hb_178817_010 |
0.2460853609 |
- |
- |
protease [Gossypium hirsutum] |
| 11 |
Hb_003947_010 |
0.2497624445 |
- |
- |
hypothetical protein JCGZ_24829 [Jatropha curcas] |
| 12 |
Hb_001140_340 |
0.2525035525 |
- |
- |
- |
| 13 |
Hb_083270_010 |
0.2600705874 |
- |
- |
hypothetical protein ARALYDRAFT_896220 [Arabidopsis lyrata subsp. lyrata] |
| 14 |
Hb_006198_170 |
0.2625695047 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 15 |
Hb_000045_270 |
0.2685673504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010452_030 |
0.2685690085 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 17 |
Hb_001100_020 |
0.2685789376 |
- |
- |
- |
| 18 |
Hb_122911_010 |
0.2686460124 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 19 |
Hb_002400_110 |
0.2686760535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_072149_010 |
0.2686828562 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |