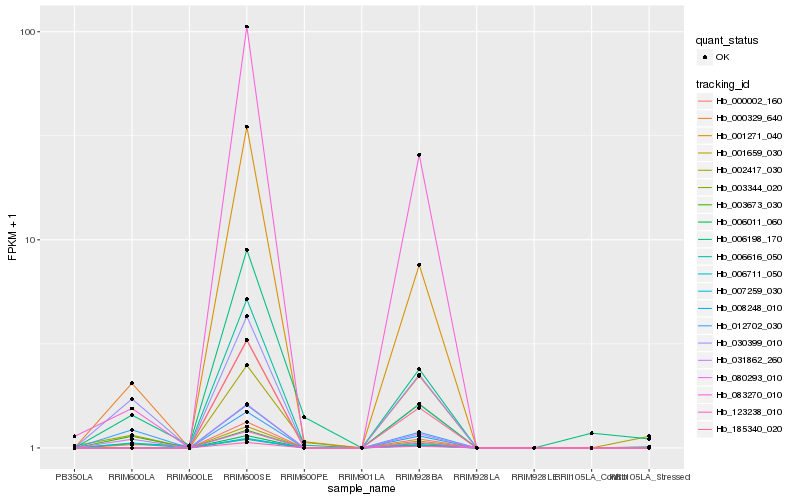
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_260 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_030399_010 |
0.0656484227 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 3 |
Hb_012702_030 |
0.157924742 |
- |
- |
hypothetical protein POPTR_0077s002001g, partial [Populus trichocarpa] |
| 4 |
Hb_006711_050 |
0.1704081702 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 5 |
Hb_000329_640 |
0.1763166942 |
- |
- |
- |
| 6 |
Hb_083270_010 |
0.2222764382 |
- |
- |
hypothetical protein ARALYDRAFT_896220 [Arabidopsis lyrata subsp. lyrata] |
| 7 |
Hb_006616_050 |
0.2259916383 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 8 |
Hb_006198_170 |
0.2261525786 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 9 |
Hb_003673_030 |
0.2311479761 |
- |
- |
hypothetical protein POPTR_0011s11940g [Populus trichocarpa] |
| 10 |
Hb_002417_030 |
0.2381527414 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 11 |
Hb_123238_010 |
0.2445494053 |
- |
- |
hypothetical protein JCGZ_16598 [Jatropha curcas] |
| 12 |
Hb_003344_020 |
0.2446635366 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103501197 [Cucumis melo] |
| 13 |
Hb_001659_030 |
0.2447551921 |
- |
- |
- |
| 14 |
Hb_007259_030 |
0.2447576772 |
- |
- |
- |
| 15 |
Hb_185340_020 |
0.2447765211 |
- |
- |
- |
| 16 |
Hb_080293_010 |
0.2451102446 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Nelumbo nucifera] |
| 17 |
Hb_001271_040 |
0.2451132159 |
- |
- |
hypothetical protein POPTR_0019s039903g, partial [Populus trichocarpa] |
| 18 |
Hb_008248_010 |
0.2454183014 |
- |
- |
- |
| 19 |
Hb_000002_160 |
0.2455100556 |
- |
- |
Uncharacterized protein TCM_021030 [Theobroma cacao] |
| 20 |
Hb_006011_060 |
0.2462947324 |
- |
- |
- |