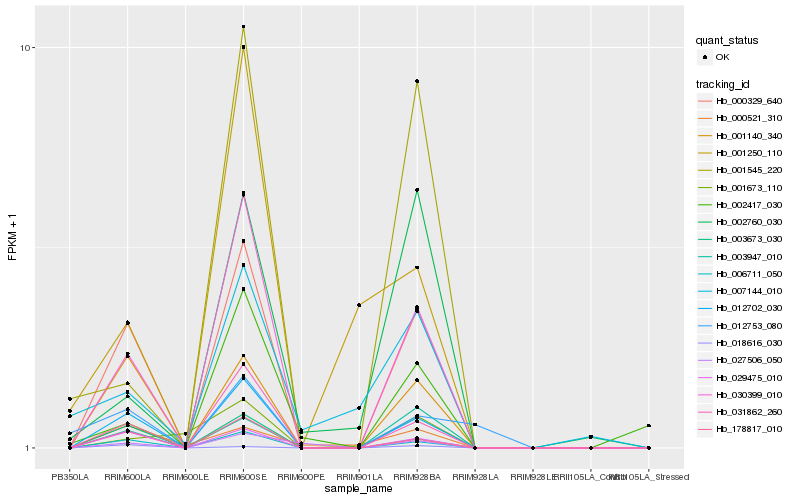
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_640 |
0.0 |
- |
- |
- |
| 2 |
Hb_012702_030 |
0.0576540874 |
- |
- |
hypothetical protein POPTR_0077s002001g, partial [Populus trichocarpa] |
| 3 |
Hb_006711_050 |
0.0711069344 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 4 |
Hb_030399_010 |
0.1189611802 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 5 |
Hb_029475_010 |
0.1203717816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001140_340 |
0.1378084943 |
- |
- |
- |
| 7 |
Hb_003673_030 |
0.1576076272 |
- |
- |
hypothetical protein POPTR_0011s11940g [Populus trichocarpa] |
| 8 |
Hb_031862_260 |
0.1763166942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_178817_010 |
0.1773323282 |
- |
- |
protease [Gossypium hirsutum] |
| 10 |
Hb_003947_010 |
0.1820669647 |
- |
- |
hypothetical protein JCGZ_24829 [Jatropha curcas] |
| 11 |
Hb_027506_050 |
0.2809032191 |
- |
- |
- |
| 12 |
Hb_000521_310 |
0.2863648115 |
- |
- |
- |
| 13 |
Hb_018616_030 |
0.2897530916 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 14 |
Hb_002417_030 |
0.2926653097 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 15 |
Hb_001545_220 |
0.2972070628 |
- |
- |
hypothetical protein POPTR_0002s06170g [Populus trichocarpa] |
| 16 |
Hb_002760_030 |
0.2994796642 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 17 |
Hb_001673_110 |
0.3054871675 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 18 |
Hb_007144_010 |
0.3216682025 |
- |
- |
- |
| 19 |
Hb_012753_080 |
0.3270389381 |
- |
- |
- |
| 20 |
Hb_001250_110 |
0.3302323684 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0010s05340g [Populus trichocarpa] |