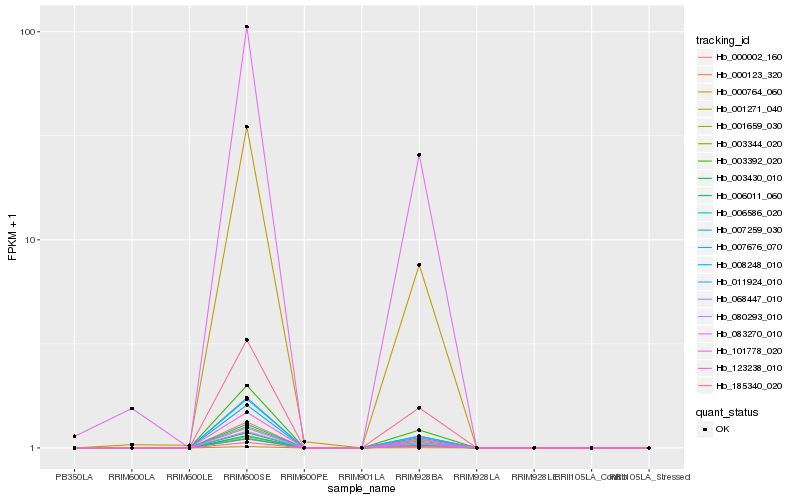
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083270_010 |
0.0 |
- |
- |
hypothetical protein ARALYDRAFT_896220 [Arabidopsis lyrata subsp. lyrata] |
| 2 |
Hb_008248_010 |
0.0513836606 |
- |
- |
- |
| 3 |
Hb_185340_020 |
0.0516103569 |
- |
- |
- |
| 4 |
Hb_007259_030 |
0.0516406964 |
- |
- |
- |
| 5 |
Hb_006011_060 |
0.0523393899 |
- |
- |
- |
| 6 |
Hb_003392_020 |
0.0524773112 |
- |
- |
PREDICTED: uncharacterized protein LOC105650609 [Jatropha curcas] |
| 7 |
Hb_003430_010 |
0.0542246942 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 33 precursor [Populus trichocarpa] |
| 8 |
Hb_000123_320 |
0.0547268229 |
- |
- |
PREDICTED: MADS-box protein FLOWERING LOCUS C-like [Jatropha curcas] |
| 9 |
Hb_006586_020 |
0.0576831301 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_001271_040 |
0.0597291602 |
- |
- |
hypothetical protein POPTR_0019s039903g, partial [Populus trichocarpa] |
| 11 |
Hb_101778_020 |
0.0597594659 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_123238_010 |
0.0629917343 |
- |
- |
hypothetical protein JCGZ_16598 [Jatropha curcas] |
| 13 |
Hb_011924_010 |
0.0635572197 |
- |
- |
hypothetical protein, partial [Phaeodactylibacter xiamenensis] |
| 14 |
Hb_003344_020 |
0.0641828533 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103501197 [Cucumis melo] |
| 15 |
Hb_001659_030 |
0.0650638537 |
- |
- |
- |
| 16 |
Hb_080293_010 |
0.068073947 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Nelumbo nucifera] |
| 17 |
Hb_000002_160 |
0.0709932894 |
- |
- |
Uncharacterized protein TCM_021030 [Theobroma cacao] |
| 18 |
Hb_068447_010 |
0.0717618677 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_007676_070 |
0.0743863939 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1-like [Populus euphratica] |
| 20 |
Hb_000764_060 |
0.0754505827 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |