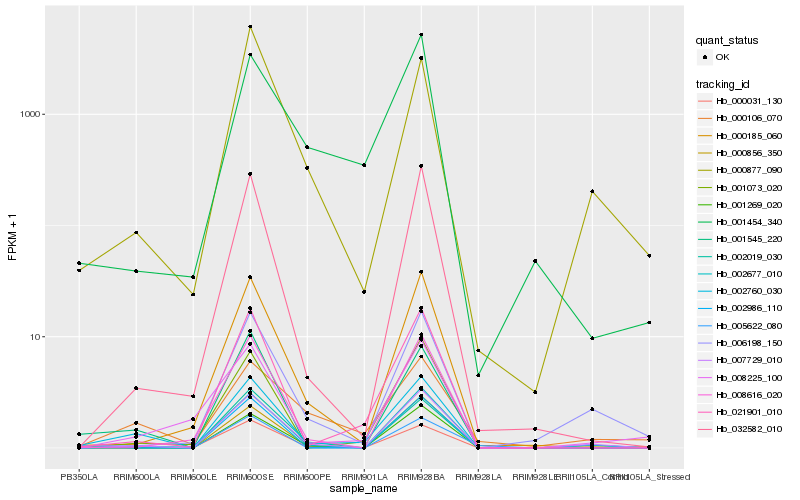
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002760_030 |
0.0 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 2 |
Hb_007729_010 |
0.1551102283 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_021901_010 |
0.1636023276 |
- |
- |
- |
| 4 |
Hb_001545_220 |
0.1667155052 |
- |
- |
hypothetical protein POPTR_0002s06170g [Populus trichocarpa] |
| 5 |
Hb_001269_020 |
0.179096602 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 6 |
Hb_002677_010 |
0.1799783694 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 7 |
Hb_001073_020 |
0.1837422527 |
- |
- |
hypothetical protein CISIN_1g031854mg [Citrus sinensis] |
| 8 |
Hb_032582_010 |
0.1870782957 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 9 |
Hb_000185_060 |
0.1965688304 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002019_030 |
0.1986266564 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_000856_350 |
0.2000283403 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 36 [Jatropha curcas] |
| 12 |
Hb_000106_070 |
0.2015672214 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 13 |
Hb_006198_150 |
0.2021731797 |
- |
- |
- |
| 14 |
Hb_001454_340 |
0.2028702467 |
- |
- |
ATDI21 [Manihot esculenta] |
| 15 |
Hb_008616_020 |
0.2035613188 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000031_130 |
0.203648948 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 17 |
Hb_008225_100 |
0.2049562503 |
- |
- |
- |
| 18 |
Hb_000877_090 |
0.2058327177 |
- |
- |
allergenic-related protein Pt2L4 [Manihot esculenta] |
| 19 |
Hb_005622_080 |
0.2061090606 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 20 |
Hb_002986_110 |
0.2069030326 |
- |
- |
PREDICTED: patatin-like protein 1 [Jatropha curcas] |