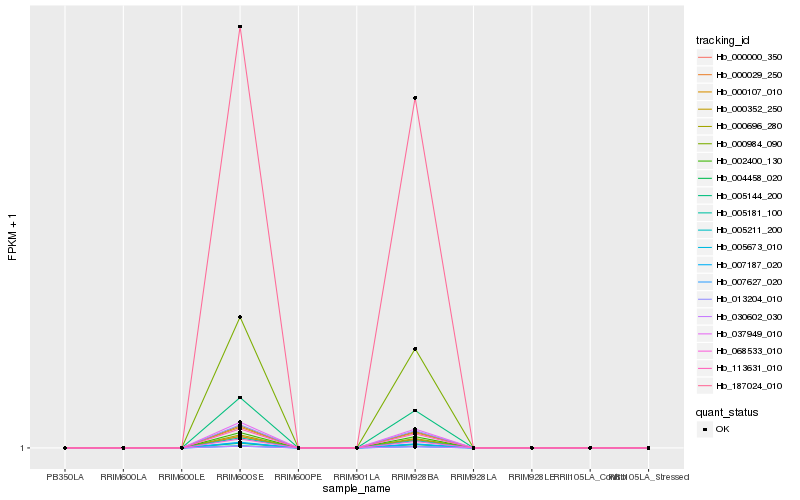
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005211_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 2 |
Hb_004458_020 |
0.0001119436 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 3 |
Hb_007187_020 |
0.0001525765 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 4 |
Hb_068533_010 |
0.0001953448 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000696_280 |
0.0004118432 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 6 |
Hb_005673_010 |
0.000832635 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_030602_030 |
0.0009639632 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_007627_020 |
0.0010099737 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_005181_100 |
0.001467979 |
- |
- |
hypothetical protein POPTR_0010s15550g [Populus trichocarpa] |
| 10 |
Hb_000000_350 |
0.0018744769 |
- |
- |
- |
| 11 |
Hb_000107_010 |
0.0019251163 |
- |
- |
- |
| 12 |
Hb_037949_010 |
0.0023568777 |
- |
- |
hypothetical protein JCGZ_11175 [Jatropha curcas] |
| 13 |
Hb_000984_090 |
0.0025252448 |
- |
- |
- |
| 14 |
Hb_002400_130 |
0.0030530932 |
- |
- |
PREDICTED: uncharacterized protein LOC105637626 [Jatropha curcas] |
| 15 |
Hb_005144_200 |
0.003714327 |
- |
- |
PREDICTED: uncharacterized protein LOC105642066 [Jatropha curcas] |
| 16 |
Hb_113631_010 |
0.0041908278 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_000029_250 |
0.0047060137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000352_250 |
0.0051243008 |
- |
- |
- |
| 19 |
Hb_013204_010 |
0.0054586299 |
- |
- |
hypothetical protein CICLE_v10006597mg [Citrus clementina] |
| 20 |
Hb_187024_010 |
0.0090999783 |
- |
- |
- |