| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
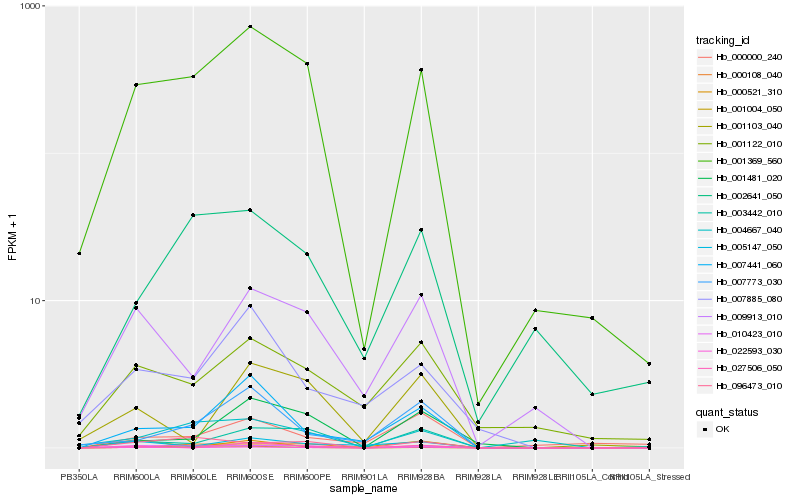
Hb_022593_030 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_001369_560 |
0.1447601404 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 3 |
Hb_010423_010 |
0.1486041294 |
- |
- |
- |
| 4 |
Hb_003442_010 |
0.2027725202 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 5 |
Hb_009913_010 |
0.2258413958 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_007441_060 |
0.232792889 |
- |
- |
- |
| 7 |
Hb_005147_050 |
0.2413462222 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 8 |
Hb_096473_010 |
0.250407417 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_001122_010 |
0.2561016972 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_007885_080 |
0.268167683 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001481_020 |
0.2713128605 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 12 |
Hb_007773_030 |
0.2742336648 |
- |
- |
- |
| 13 |
Hb_000521_310 |
0.2758822318 |
- |
- |
- |
| 14 |
Hb_027506_050 |
0.2767363862 |
- |
- |
- |
| 15 |
Hb_000108_040 |
0.2789280412 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Gossypium raimondii] |
| 16 |
Hb_001004_050 |
0.2813278873 |
- |
- |
PREDICTED: uncharacterized protein LOC103335231 [Prunus mume] |
| 17 |
Hb_001103_040 |
0.2832588117 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 18 |
Hb_002641_050 |
0.2867227024 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 19 |
Hb_000000_240 |
0.2875430439 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 20 |
Hb_004667_040 |
0.2885781255 |
- |
- |
ATP binding protein, putative [Ricinus communis] |