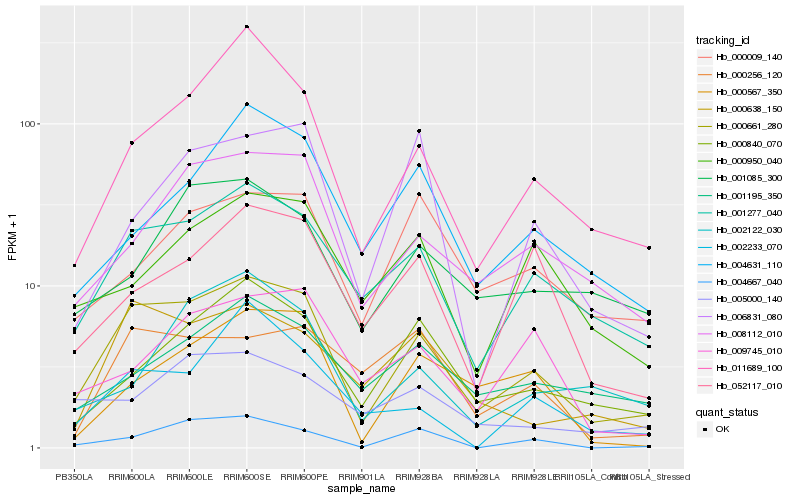
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_280 |
0.0 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 2 |
Hb_001277_040 |
0.1339772149 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 3 |
Hb_008112_010 |
0.1720306272 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 4 |
Hb_011689_100 |
0.173317588 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 5 |
Hb_000638_150 |
0.1816595692 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 6 |
Hb_000840_070 |
0.1884755038 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_004667_040 |
0.1958944873 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_001195_350 |
0.2024303663 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 9 |
Hb_001085_300 |
0.2032713739 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 10 |
Hb_052117_010 |
0.2059925145 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_004631_110 |
0.2069826552 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_000256_120 |
0.2074873637 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 13 |
Hb_005000_140 |
0.2092027584 |
- |
- |
PREDICTED: uncharacterized protein LOC105637876 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000567_350 |
0.2103534062 |
- |
- |
PREDICTED: probable acyl-activating enzyme 6 [Jatropha curcas] |
| 15 |
Hb_002233_070 |
0.2105152318 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000009_140 |
0.2108566726 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 17 |
Hb_000950_040 |
0.2108893237 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 18 |
Hb_009745_010 |
0.211472756 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 19 |
Hb_006831_080 |
0.2132553378 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 20 |
Hb_002122_030 |
0.2158747879 |
- |
- |
- |