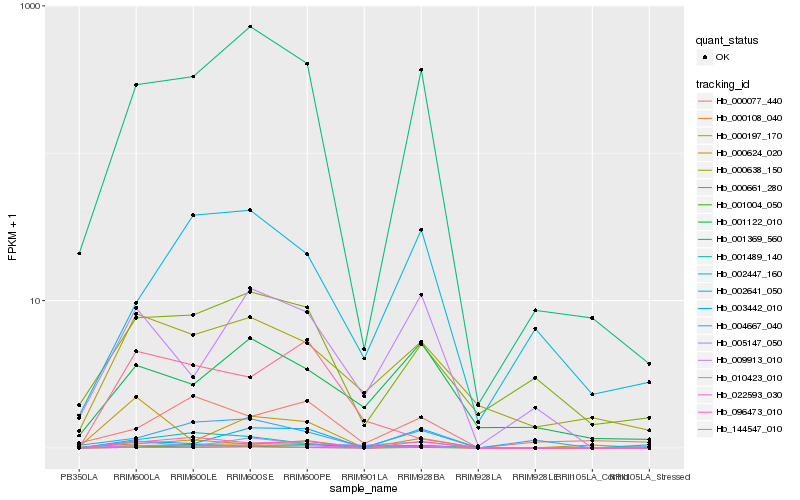
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010423_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_022593_030 |
0.1486041294 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 3 |
Hb_001369_560 |
0.1616602855 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 4 |
Hb_096473_010 |
0.1620217887 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_000108_040 |
0.2434355993 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Gossypium raimondii] |
| 6 |
Hb_003442_010 |
0.2525577723 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 7 |
Hb_001004_050 |
0.2599908743 |
- |
- |
PREDICTED: uncharacterized protein LOC103335231 [Prunus mume] |
| 8 |
Hb_009913_010 |
0.2647350371 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000638_150 |
0.2651462941 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 10 |
Hb_000197_170 |
0.2683295491 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000661_280 |
0.2686922186 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 12 |
Hb_005147_050 |
0.2884349953 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 13 |
Hb_001489_140 |
0.2893277927 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 14 |
Hb_004667_040 |
0.2912400857 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_001122_010 |
0.2932188825 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 16 |
Hb_002641_050 |
0.3038124328 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 17 |
Hb_000624_020 |
0.3056478745 |
transcription factor |
TF Family: NAC |
NAC transcription factor 004 [Jatropha curcas] |
| 18 |
Hb_002447_160 |
0.3082730747 |
- |
- |
- |
| 19 |
Hb_000077_440 |
0.3094265799 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 20 |
Hb_144547_010 |
0.3099042181 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |