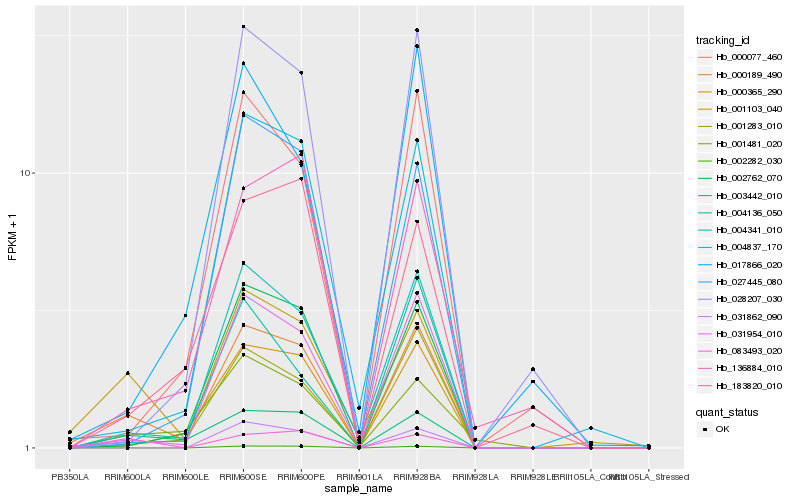
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_490 |
0.0 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 2 |
Hb_031954_010 |
0.1410197347 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001103_040 |
0.1416712053 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 4 |
Hb_003442_010 |
0.1442675138 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 5 |
Hb_004341_010 |
0.1667878291 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 6 |
Hb_001283_010 |
0.1672857364 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 7 |
Hb_000365_290 |
0.1684748037 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002762_070 |
0.1728095942 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 9 |
Hb_004837_170 |
0.1812011322 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 10 |
Hb_001481_020 |
0.1833526864 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 11 |
Hb_000077_460 |
0.1855655769 |
- |
- |
- |
| 12 |
Hb_004136_050 |
0.1883976055 |
- |
- |
- |
| 13 |
Hb_028207_030 |
0.1934597342 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 14 |
Hb_136884_010 |
0.1955955142 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 15 |
Hb_027445_080 |
0.1969355681 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 16 |
Hb_183820_010 |
0.2002085081 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 17 |
Hb_083493_020 |
0.2067735891 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Vitis vinifera] |
| 18 |
Hb_002282_030 |
0.2093412792 |
- |
- |
- |
| 19 |
Hb_031862_090 |
0.2097736075 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 20 |
Hb_017866_020 |
0.2105366991 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |