| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
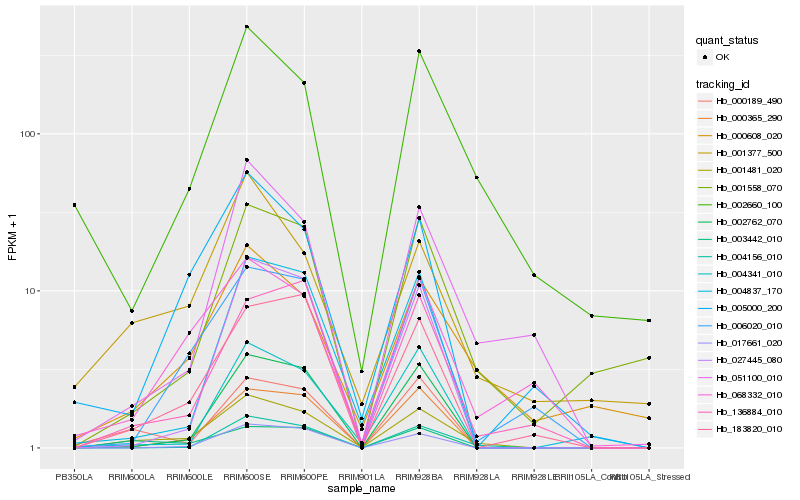
Hb_001481_020 |
0.0 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 2 |
Hb_003442_010 |
0.1672214849 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 3 |
Hb_000608_020 |
0.1788982164 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 4 |
Hb_000189_490 |
0.1833526864 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 5 |
Hb_000365_290 |
0.1837159094 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004156_010 |
0.1900536284 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 7 |
Hb_068332_010 |
0.1920327227 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_002762_070 |
0.1926420588 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 9 |
Hb_051100_010 |
0.1950721464 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_006020_010 |
0.1961616377 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 11 |
Hb_183820_010 |
0.1976420618 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 12 |
Hb_001377_500 |
0.1976453433 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 13 |
Hb_002660_100 |
0.2002951765 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 14 |
Hb_005000_200 |
0.2038042571 |
- |
- |
PREDICTED: uncharacterized protein LOC105637887 [Jatropha curcas] |
| 15 |
Hb_136884_010 |
0.2043721237 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_001558_070 |
0.2051235867 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 17 |
Hb_017661_020 |
0.2071364635 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 18 |
Hb_004837_170 |
0.207338779 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 19 |
Hb_027445_080 |
0.2076351558 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 20 |
Hb_004341_010 |
0.2089367648 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |